AbstractPurpose Brain metastasis rarely occurs in soft tissue sarcoma (STS). Here, we present five cases of STS with brain metastases with genetic profiles.
Materials and Methods We included five patients from Seoul National University Hospital who were diagnosed with STS with metastasis to the brain. Tissue from the brain metastasis along with that from the primary site or other metastases were used for DNA and RNA sequencing to identify genetic profiles. Gene expression profiles were compared with sarcoma samples from The Cancer Genome Atlas.
Results The overall survival after diagnosis of brain metastasis ranged from 2.2 to 34.3 months. Comparison of mutational profiles between brain metastases and matched primary or other metastatic samples showed similar profiles. In two patients, copy number variation profiles between brain metastasis and other tumors showed several differences including MYCL, JUN, MYC, and DDR2 amplification. Gene ontology analysis showed that the group of genes significantly highly expressed in the brain metastasis samples was enriched in the G-protein coupled receptor activity, structural constituent of chromatin, protein heterodimerization activity, and binding of DNA, RNA, and protein. Gene set enrichment analysis showed enrichment in the pathway of neuroactive ligand-receptor interaction and systemic lupus erythematosus.
IntroductionSarcomas are a heterogeneous group of diseases that account for approximately 1% of cancers worldwide [1,2]. Multimodality treatment with surgery, radiotherapy, and chemotherapy is used to treat most sarcomas [2]. However, metastatic sarcoma is a major challenge that limits patient survival. While the common sites of sarcoma metastasis include the lungs and liver, sarcomas can metastasize to the brain [3-5].
Brain metastasis of sarcomas shows an incidence of around 1%-8% of patients with sarcomas, which is rare considering the incidence of sarcomas and the previous report that 10%-30% of cancer patients in general develop brain metastasis [4]. However, the incidence of brain metastasis of sarcomas is expected to rise as there are developing new technologies for detecting metastasis and for improving the survival of sarcomas through evolving treatment strategies [3,6,7]. A previous study on 112 cases of brain metastasis of sarcoma over 28 years showed that various types of sarcomas can metastasize to the brain with undifferentiated sarcoma being the most common followed by alveolar soft part sarcoma and osteosarcoma [3]. Brain metastasis of sarcoma is treated with multiple modalities, although the treatment objective is often palliative [5]. Therefore, the prognosis of sarcoma brain metastasis is often dismal, with a majority of patients surviving less than 12 months [5]. However, there is also a small population with a longer survival time after the aggressive control of brain metastasis [5]. This observation prompts further investigation into the biology of brain metastasis of sarcoma to search for additional treatment options.
Although there is no universally applicable mechanism that promotes brain metastasis across all types of cancers, several genomic and transcriptomic analyses have been reported which demonstrate the potential mechanism in other cancer types. Genomic analysis of matched brain metastases and primary tumors, mostly from the lung, breast, and kidney, demonstrate that 53 % of cases harbor additional pathogenic alterations in the brain metastases that are not present in the primary tumors, such as PTEN loss, PIK3CA mutation, and BRCA2 mutation [8]. Gene expression analysis of resected melanoma and matched extracranial metastases identified significant suppression of immune cell networks and the enrichment of oxidative phosphorylation, which was associated with resistance to therapeutics for melanoma [9]. Transcriptomic analysis of the brain metastasis of breast cancer showed acquisition of RET expression and human epidermal growth receptor 2 signaling enhancement, which could have provided additional proliferative advantages [10]. Case-control analysis of lung adenocarcinoma, with or without brain metastases, showed a higher frequency of gene copy number variation, including MYC amplification, YAP1 amplification, and CDKN2A/B deletion [11]. We hypothesized that such mutations, copy number variations, and expression changes could be observed in the brain metastases of sarcomas.
Here, we present five cases of sarcoma and brain metastases. We performed genomic and transcriptomic sequencing of brain metastases and their matched primary and/or other metastatic tumors. Through these analyses, we searched the potential genetic mechanisms that might promote brain metastasis of sarcomas.
Materials and Methods1. Patients and specimen collectionThe study cohort consisted of patients at Seoul National University Hospital who were diagnosed with soft tissue sarcoma with metastasis to the brain. In addition, the tissue from the brain metastasis along with that from the primary site or other metastases needed to be available for sequencing. Five patients met these criteria. All tissues from these patients were stored as formalin-fixed paraffin-embedded tissues collected during the clinical process of treating these patients outside of the study setting.
The demographics, histological characteristics, radiographic findings, treatments, and outcomes of these patients were retrieved retrospectively from electronic medical records. All data collection, sequencing, and analyses were performed with the approval of the Institutional Review Board of Seoul National University Hospital (IRB No. 2110-120-1263) and in accordance with the Declaration of Helsinki. All patients in this study provided informed consents on the donation of their human biological materials for purposes of research.
2. Sequencing data analysisDetailed methods on the sequencing data analysis are available in Supplementary Methods. For exome sequencing analysis, ‘Oncogenic’ and ‘Likely Oncogenic’ mutations by OncoKB-Annotator were identified as somatic mutations [12]. When generating the figure for genomic analyses, a copy number variation heatmap was created using Integrative Genomics Viewer (2.8.9) [13]. The scale of the heatmap was adjusted according to the copy number, and mutation types were drawn online using OncoPrinter [14,15]. Above analysis was carried out using the computing server at the Genomic Medicine Institute Research Service Center. These sequence data have been submitted to the GenBank Sequence Read Archive under the accession numbers PRJNA922237. Sample quality control results are available in S1 Table.
3. Transcriptomics analysisTo compare the transcriptomic data of the study cohort with the transcriptomic data of The Cancer Genome Atlas (TCGA) sarcoma (SARC) [1], we used the read count data and performed normalization using the trimmed mean of M values algorithm [16]. The read count data of TCGA-SARC was downloaded from the Xena platform [17]. The aggressive fibromatosis cases in the TCGA-SARC cohort were excluded, and the remaining 263 cases were included for the analysis.
To perform the gene ontology analysis, we first calculated the fold changes of the means and p-values by Student’s t test comparing the gene expression values of the study cohort to TCGA-SARC. We selected genes with p-values less than 1.0×10–7 and fold changes greater than 2.0, or less than 0.5. The group of genes that were expressed significantly higher in the study cohort (i.e., p < 1.0×10–7 and fold changes > 2.0) was used as the list for the gene ontology analysis using the molecular function gene ontology term provided by the Database for Annotation, Visualization and Integrated Discovery ver. 6.8 [18]. A p-value less than 0.05 in the gene ontology analysis was considered as a significantly enriched gene ontology term.
Next, we evaluated the gene expression pathways that were enriched in the study cohort using Gene Set Enrichment Analysis (GSEA) software ver. 4.1 [19]. The gene sets used for the GSEA were from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways [20]. A nominal p-value of less than 0.05 was considered a significantly enriched pathway.
Results1. Patient characteristicsA total of 11 samples from the five patients including brain metastases, primary tumors, and other metastatic tumors were available for DNA sequencing and RNA sequencing, respectively. Detailed information on the demographics of the five patients and the sequenced sites is available in Table 1. The details of the medical histories of these patients are as follows.
(1) Patient AA 49-year-old male complained of dyspnea and was diagnosed with a pericardial tumor. The mass was resected to reveal synovial sarcoma, and the patient was treated with six cycles of ifosfamide and doxorubicin. One year after the chemotherapy, the disease relapsed in the mediastinum, and the patient received second-line pazopanib followed by a third-line combination of gemcitabine and dacarbazine, to which the disease eventually progressed. The patient then received palliative radiotherapy for the mass around the superior vena cava and heart, during which the patient complained of acute visual disturbance. Brain imaging showed an intracerebral hemorrhage around a 1-cm-sized enhancing lesion in the right temporo-occipital lobe, which was surgically removed. Tissue pathology showed the same synovial sarcoma. However, the patient deteriorated and died 2.2 months after the discovery of brain metastasis.
(2) Patient BA 15-year-old female complained of a resected mass in the left thigh that was found to have synovial sarcoma. The patient received adjuvant concurrent chemoradiotherapy with a combination of ifosfamide and etoposide followed by a combination of ifosfamide and doxorubicin. However, the disease relapsed in the lungs, and the patient underwent lung metastasectomy along with four lines of combination chemotherapy, including pazopanib and radiotherapy. However, the patient complained of headache and visual field disturbance, for which brain imaging showed a 6.3-cm-sized hemorrhagic metastasis in the left parieto-occipital lobe with perilesional edema. The tumor was surgically removed, and the pathology returned to the same synovial sarcoma. The patient deteriorated and died 2.8 months after the discovery of brain metastasis.
(3) Patient CA 57-year-old male complained of a right upper back mass, the excisional biopsy of which showed undifferentiated pleomorphic sarcoma. The mass was excised. After 4 years and three months, the disease relapsed with lung metastasis, which was treated with lung metastasectomy, adjuvant ifosfamide, and doxorubicin. However, the patient complained of monoparesis of the arm after 6 months, and brain imaging showed a 4.3-cm-sized multicystic and solid enhancing mass with hemorrhage in the right frontal lobe accompanied by perilesional edema. The tumor was resected, and the patient was alive and followed up for 30.0 months after the discovery of brain metastasis without disease relapse.
(4) Patient DA 42-year-old female presented with a mass on the left gluteus. A wide excision was performed, and the mass was found to be a high-grade pleomorphic liposarcoma. The patient underwent adjuvant radiotherapy at the primary site. However, there was an immediate recurrence with multiple lung metastases, for which the patient received two lines of chemotherapy, including immune checkpoint inhibitors. However, the patient complained of left-sided hemiparesis. Brain imaging showed enhancing masses in the right frontoparietal and left occipital lobes, and the patient underwent resection of the brain tumor, followed by gamma knife surgery. Pathology of the brain mass showed pleomorphic liposarcoma. After 8 months, the disease had metastasized to the small bowel, which was resected, followed by eribulin. The disease remained stable, and the patient is currently alive 17.5 months after the discovery of brain metastasis.
(5) Patient EA 35-year-old male presented with headache and dysarthria that started 1 month prior to presentation. The work-up of the mass showed alveolar soft part sarcoma with multiple metastases to the brain, lungs, and bones. The patient underwent brain tumor resection, followed by gamma knife surgery. Subsequently, the patient underwent multiple rounds of radiotherapy and palliative chemotherapy with doxorubicin combined with olaratumab. However, the disease progressed despite the chemotherapy, and the performance status of the patient rapidly deteriorated. As a result, the patient did not undergo any further systemic treatment. The patient died 34.3 months after the discovery of brain metastasis.
2. Genomic analysisA summary of the pathogenic variants detected in these samples is presented in Table 2. TP53 mutation was detected in two cases. Otherwise, the cases did not show a mutation of a gene in common. Comparison of mutational profiles between brain metastasis and matched primary or other metastasis samples showed identical profiles with only subtle differences in allele frequency values.
Patients C and D showed several differences in patterns of copy number variation profiles between brain metastasis and the primary and other metastatic tumors (Fig. 1, S2 Table). In the case of patient C, while DDR2, NTRK1, and MCL1 amplification and RB1 deletion were observed in the primary and brain metastatic tumors, JUN, MYC, and MYCL amplification were found only in the brain metastasis. In patient D, PIK3CA, NTRK1, and MCL1 amplification was detected in the metastatic tumors, but not in the primary tumor. In addition, DDR2 amplification was observed only in brain metastases. PTEN and FAS deletions were observed in primary and metastatic tumors.
Summary on the structural variant is available in Table 3. No significant structural variant changes were observed between matched samples.
3. Expression analysisIn addition to genomic evolution from the genomic data, we hypothesized that the samples from brain metastases would share similar characteristics with matched primary and metastatic tumors. Therefore, we performed differential gene expression analysis between the 11 samples from the five patients with brain metastasis and TCGA-SARC samples (n=263). After the selection process described above, we found genes that were expressed significantly differently between the groups (Fig. 2A). Gene ontology analysis showed that the group of genes that were significantly highly expressed in the brain metastasis samples were enriched in G-protein coupled receptor activity, olfactory receptor activity, taste receptor activities, structural constituents of chromatin, protein heterodimerization activity, and DNA binding (Fig. 2A). GSEA showed that the brain metastasis samples were enriched in the olfactory transduction pathway, neuroactive ligand-receptor interaction, systemic lupus erythematosus, and taste transduction (Fig. 2B and C).
DiscussionIn this study, we presented the clinical courses of five patients with brain metastases of sarcoma with analysis of the genomic and transcriptomic profiles of these tumors with matched primary or extracranial metastases. Two patients died within 3 months after the diagnosis of brain metastases. However, three patients, including one with brain metastases at the time of sarcoma diagnosis, survived more than 12 months after diagnosis of brain metastases with aggressive treatment of brain metastases through surgical removal and gamma knife surgery. Our observations suggest that some patients would benefit from aggressive treatment, as implicated in previous studies [5]. Therefore, investigation is needed to evaluate the potential mechanism of brain metastases of sarcomas to better understand its characteristics.
In the sequencing data, the samples from the five patients harbored well-known genomic alterations commonly observed in the previous literature. The two samples from synovial sarcoma showed SS18-SSX1 fusion [1]. The sample from undifferentiated pleomorphic sarcoma showed TP53 mutation, ATRX mutation, RB1 loss, and high copy number amplifications [1,21]. The pleomorphic liposarcoma sample showed TP53 mutation and loss-of-function RB1 mutation [21,22]. Finally, the sample from alveolar soft part sarcoma showed ASPSR1-TFE3 fusion [23]. These results suggest that sarcomas that metastasize to brain do not necessarily vary significantly from sarcomas without brain metastasis.
When comparing the brain metastases with other matched tumors, there were no additional oncogenic mutations in brain metastases compared with their matched other tumors. There were two patients with copy number variation changes in brain metastases. This genomic pattern is different compared to lung, breast, and kidney cancers, which show frequent additional driver mutations in brain metastases [8], in that the oncogenic mutational profiles between brain metastases and extracranial tumors were similar [24]. Also, previous study evaluating genomic patterns between primary and metastatic tumors showed that there were no significant genomic changes in sarcomas [25]. Our findings may not be surprising considering that sarcomas have a very low mutational burden and that acquisition of driver mutations are not mandatory for sarcoma progression [1]. Additionally, previous studies on the clonal evolution of sarcoma have found a paucity of clonal evolution at the DNA level, suggesting that a sarcoma obtains full genetic optimization early in tumorigenesis [26].
We observed differential expression of genes in sarcomas that metastasized to the brain compared to sarcomas without brain metastasis. The expression profile modification of brain metastases compared to primary tumors has already been demonstrated in melanoma, breast cancer, and lung cancer [9-11]. The pathway that was elucidated in both the gene ontology and GSEA analysis was epigenetic modification, which includes the histone pathway termed as ‘systemic lupus erythematosus’ in the KEGG pathway used for GSEA analysis [20]. Epigenetic modifications have been widely discussed in various types of cancers. Specifically, a previous study reported that increased histone expression is present in breast cancer and is associated with poor survival in breast cancer patients [27]. Other pathways identified in our study were associated with tumor microenvironment and metastasis in previous literature. For example, the neuroactive ligand-receptor interaction pathway helped tumors to adapt to the target organ environment in brain and liver metastases across several types of cancers [28]. G-protein coupled receptors such as chemokine receptors that affect tumor microenvironments facilitated metastasis to other organs including liver, lung, and brain [29]. Previous study showed that expression signature related to mitosis and chromosomal instability may determine metastatic outcome in sarcoma [30]. Therefore, further study focusing on integrating our findings with the genetic profiles related to tumor microenvironment or chromosomal instability may provide more insights on the understanding and managing the brain metastasis of STS.
Although this is the first study on the sequencing analysis of brain metastasis of sarcomas with matched primary or extracranial metastasis, this study has several limitations. This study involved a small number of patients due to the rarity of sarcoma with brain metastasis and tissue available for analysis. As the patients underwent surgical removal of the brain metastases, they would have a predilection for more healthy demographics and more favorable tumor profiles. Patients with more aggressive sarcomas were not included in our study. Also, the STS subtypes of patients were heterogeneous, and the overall survival outcome was variable, which implicate that our patients may not represent the whole subtypes of patients with STS. Therefore, careful generalization of our study results is necessary. In addition, we assumed that the TCGA transcriptomic data represented general sarcomas that did not metastasize to the brain, which could cause significant selection bias. In addition, the batch effect and the differences in the tissue acquisition sites between TCGA data and the study cohort should not be overlooked, although we performed normalization to minimize the batch effect. Finally, the differentially expressed genes and pathways identified in this study need to be interpreted carefully. Our findings are from single group of patients and therefore require further validation study with experimental data and external dataset.
In this case series, patients with brain metastasis of STS show heterogeneous clinical course with variable overall survival. In these patients, no additional significant driver gene mutations were observed in the brain metastases, and two patients showed copy number variation changes. The STS with brain metastasis showed expression profile modification possibly involving epigenetic changes. Our observations suggest that evaluation of potential mechanism of brain metastasis may provide insight into selecting patients who may need more aggressive local treatment in patients with brain metastasis of STS.
Electronic Supplementary MaterialSupplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
NotesEthical Statement All data collection, sequencing, and analyses were performed with the approval of the Institutional Review Board of Seoul National University Hospital (IRB No. 2110-120-1263) and in accordance with the Declaration of Helsinki. All patients in this study provided informed consents on the donation of their human biological materials for purposes of research. Author Contributions Conceived and designed the analysis: Park C, Kim R, Kim M. Collected the data: Park C, Kim R, Kim M, Kim TM, Han I, Kim JI, Kim HS. Contributed data or analysis tools: Park C, Kim R, Choi J, Kim M, Kim TM, Kim JI, Kim HS. Performed the analysis: Park C, Kim R, Choi J, Kim M. Wrote the paper: Park C, Kim R, Choi J, Kim M, Kim TM, Han I, Kim JI, Kim HS. AcknowledgmentsThis study was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C1277), and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A1A03047972).
Fig. 1.Genetic alterations in patients C and D. (A) Copy number variation heatmap of whole chromosomes aligned by each patient’s tumor progression timeline for patients C and D. Regions with a copy number of 2 or 3 are colored white on the scale provided to the left. Indicated genes above the heatmap are the copy numbers amplified or deleted in more than two sites within one patient. The copy number for each site is also provided. Indicated genes under each heatmap are copy numbers only amplified in brain metastasis. Magnetic resonance images are shown on the right side of each heatmap. There is no image for the primary tumor of patient C as the patient had an unplanned excision of the primary tumor. (B) Mutated genes and the types of alterations in the order of cancer progression for patients C and D. PLPS, pleomorphic liposarcoma; UPS, undifferentiated pleomorphic sarcoma. 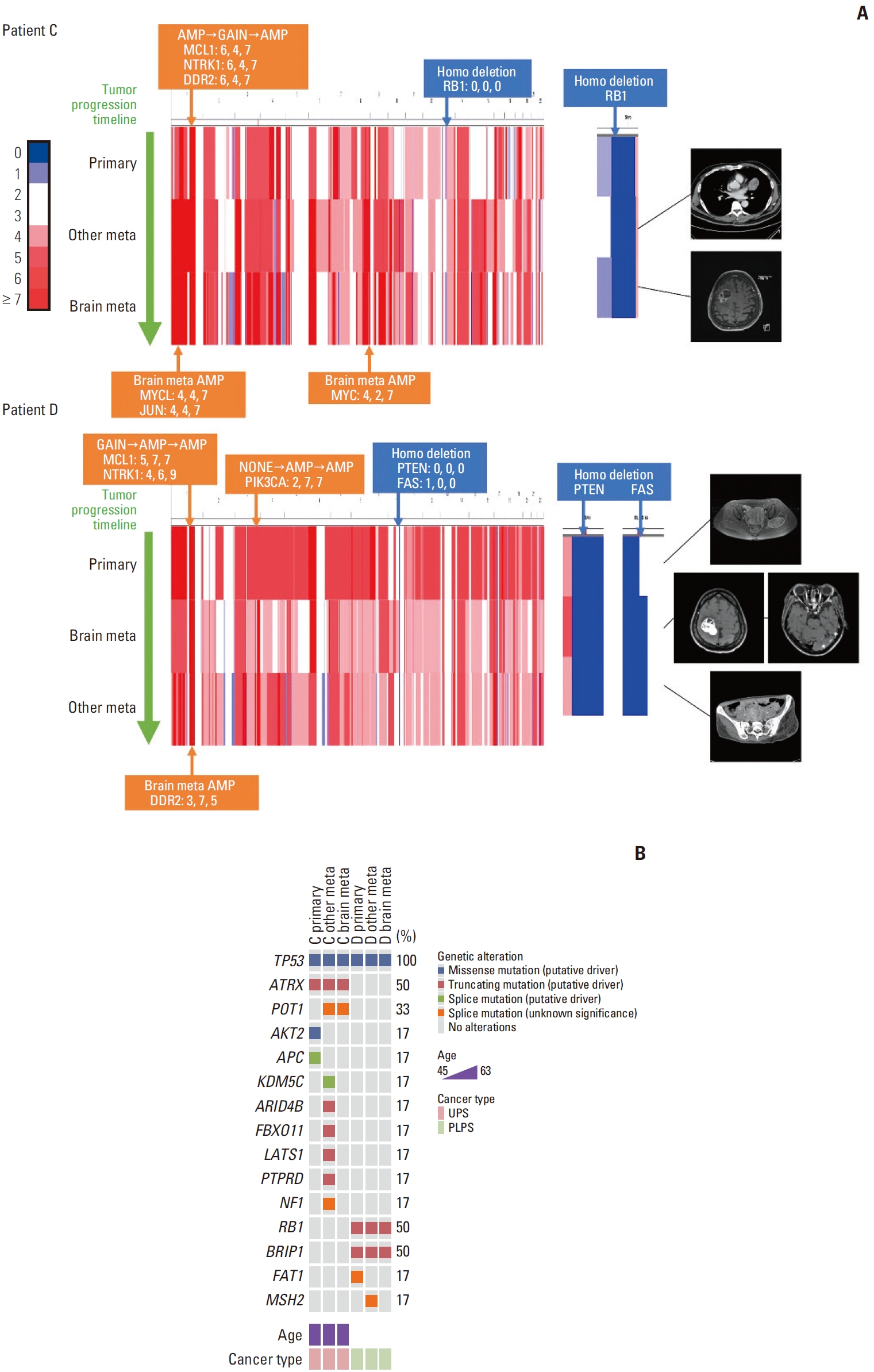
Fig. 2.Gene expression comparison of soft tissue sarcoma with brain metastasis to The Cancer Genome Atlas sarcoma (TCGA-SARC) samples. (A) The left panel shows a volcano plot. Each dot represents each gene. The x-axis value represents fold change (mean gene expression of the study cohort divided by the TCGA-SARC cohort) and the y-axis value represents the p-value. The blue dashed line denotes a fold change value of 2 and the green dashed line denotes a p-value of 1.0×10–7. Red dots are the genes that are significantly upregulated in the study cohort. Gene ontology analysis was done with the genes of the red dots. The right panel shows the significantly enriched gene ontology terms with p-values. (B) Enrichment plot of the two pathways that were significantly enriched in the study cohort by Gene Set Enrichment Analysis analysis. (C) Normalized enrichment scores of the four pathways. KEGG, Kyoto Encyclopedia of Genes and Genomes. 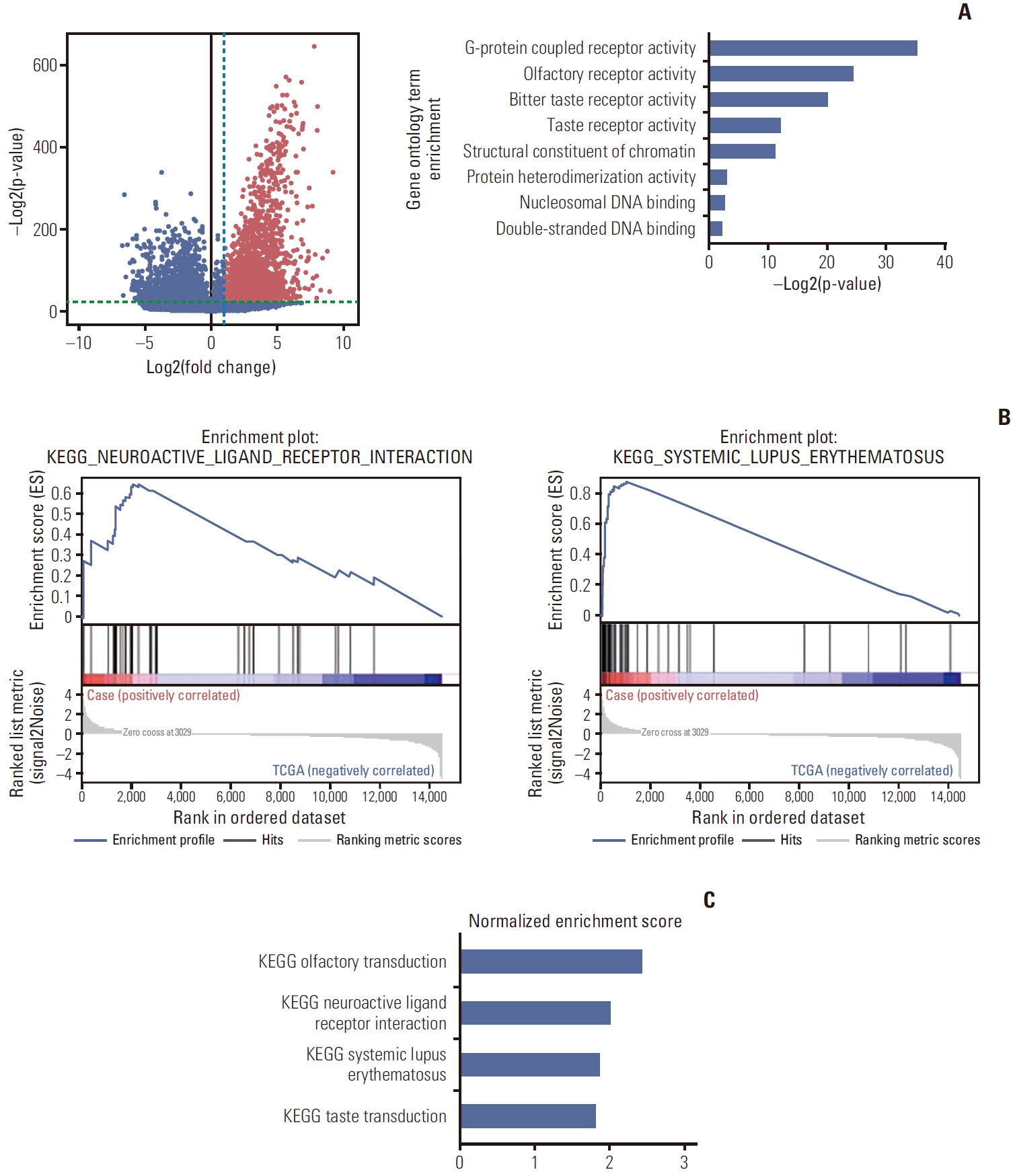
Table 1.Patient demographics Table 2.Mutational results of sequenced tumors References1. Cancer Genome Atlas Research Network. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950–96.
2. Amankwah EK, Conley AP, Reed DR. Epidemiology and therapies for metastatic sarcoma. Clin Epidemiol. 2013;5:147–62.
3. Al Sannaa G, Watson KL, Olar A, Wang WL, Fuller GN, McCutcheon I, et al. Sarcoma brain metastases: 28 years of experience at a single institution. Ann Surg Oncol. 2016;23:962–7.
4. Chaigneau L, Patrikidou A, Ray-Coquard I, Valentin T, Linassier C, Bay JO, et al. Brain metastases from adult sarcoma: prognostic factors and impact of treatment. A retrospective analysis from the French Sarcoma Group (GSF/GETO). Oncologist. 2018;23:948–55.
5. Shweikeh F, Bukavina L, Saeed K, Sarkis R, Suneja A, Sweiss F, et al. Brain metastasis in bone and soft tissue cancers: a review of incidence, interventions, and outcomes. Sarcoma. 2014;2014:475175.
6. Salvati M, D’Elia A, Frati A, Santoro A. Sarcoma metastatic to the brain: a series of 35 cases and considerations from 27 years of experience. J Neurooncol. 2010;98:373–7.
7. Gercovich FG, Luna MA, Gottlieb JA. Increased incidence of cerebral metastases in sarcoma patients with prolonged survival from chemotherapy: report of cases of leiomysarcoma and chondrosarcoma. Cancer. 1975;36:1843–51.
8. Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–77.
9. Fischer GM, Jalali A, Kircher DA, Lee WC, McQuade JL, Haydu LE, et al. Molecular profiling reveals unique immune and metabolic features of melanoma brain metastases. Cancer Discov. 2019;9:628–45.
10. Vareslija D, Priedigkeit N, Fagan A, Purcell S, Cosgrove N, O’Halloran PJ, et al. Transcriptome characterization of matched primary breast and brain metastatic tumors to detect novel actionable targets. J Natl Cancer Inst. 2019;111:388–98.
11. Shih DJ, Nayyar N, Bihun I, Dagogo-Jack I, Gill CM, Aquilanti E, et al. Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat Genet. 2020;52:371–7.
12. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017:PO.17.00011.
13. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6.
14. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
15. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
16. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25.
17. Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–8.
18. Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216–21.
19. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50.
20. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
21. Nacev BA, Sanchez-Vega F, Smith SA, Antonescu CR, Rosenbaum E, Shi H, et al. Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat Commun. 2022;13:3405.
24. Chen G, Chakravarti N, Aardalen K, Lazar AJ, Tetzlaff MT, Wubbenhorst B, et al. Molecular profiling of patient-matched brain and extracranial melanoma metastases implicates the PI3K pathway as a therapeutic target. Clin Cancer Res. 2014;20:5537–46.
25. Nguyen B, Fong C, Luthra A, Smith SA, DiNatale RG, Nandakumar S, et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell. 2022;185:563–75.
26. Hofvander J, Viklund B, Isaksson A, Brosjo O, Vult von Steyern F, Rissler P, et al. Different patterns of clonal evolution among different sarcoma subtypes followed for up to 25 years. Nat Commun. 2018;9:3662.
27. Xie W, Zhang J, Zhong P, Qin S, Zhang H, Fan X, et al. Expression and potential prognostic value of histone family gene signature in breast cancer. Exp Ther Med. 2019;18:4893–903.
28. Zhang L, Fan M, Napolitano F, Gao X, Xu Y, Li L. Transcriptomic analysis identifies organ-specific metastasis genes and pathways across different primary sites. J Transl Med. 2021;19:31.
|
|
|||||||||||||||||||||||||||||||||||||||||