AbstractPurposeBreast cancer is one of the most common causes of cancer-related death in females. Numerous drug-targetable biomarkers and predictive biomarkers have been developed. Some researchers have expressed doubts about the need for next-generation sequencing (NGS) studies in daily practice. This study analyzed the results of NGS studies on breast cancer at a single institute and evaluated the real-world applications of NGS data to precision medicine for breast cancer.
Materials and MethodsWe retrospectively collected the results of NGS studies and analyzed the histopathologic features and genetic profiles of patients treated for breast cancer from 2010 to 2021. Seventy cases had data from CancerSCAN, a customized panel of 375 cancer-associated genes, and 110 cases had data from TruSight Oncology 500.
ResultsThe most frequently detected single nucleotide variant was the TP53 mutation (123/180, 68.3%), followed by PIK3CA mutations (51/180, 28.3%). Estrogen receptor 1 (ESR1) mutation was detected in 11 patients (6.1%), of whom 10 had hormone receptor–positive, human epidermal growth factor receptor 2–negative breast cancer, and two had no history of prior endocrine therapy. Based on their NGS study results, 13 patients (7.2%) received target therapy. Among them, four patients had a BRCA1 or BRCA2 germline mutation, and nine patients had a PIK3CA mutation.
ConclusionNGS can provide information about predictive biomarkers and drug-targetable biomarkers that can enable treatment and participation in clinical trials based on precision medicine. Further studies should be conducted to excavate novel drug-targetable biomarkers and develop additional target therapies.
IntroductionBreast cancer is one of the most common malignancies and causes of cancer-related death in women including South Korea [1]. Although the development of human epidermal growth factor receptor 2 (HER2) target therapy [2,3], endocrine therapy [4], treatment for endocrine therapy resistance [5], and immune checkpoint inhibitors [6,7] has dramatically increased the survival rate of breast cancer, breast cancer patients still suffer from late recurrences after treatment and show resistance to various treatments.
The median age at diagnosis in South Korea is approximately 10 years younger (52.8 years) than that in the United States (62 years) [1,8]. In addition, the proportion of young breast cancer patients diagnosed with breast cancer under the age of 40 is higher in South Korea (10.6 %) than in the United States (5%) [1,8]. Although the prevalence of BRCA1/2 mutation in South Korea is similar to that reported in Western countries [9,10], other predisposing germline mutations in young breast cancer patients with BRCA1/2-negative were quite different in distribution [9]. As the characteristics of breast cancer in South Korea tend to be different, it is necessary to accumulate sufficient information of breast cancer in South Korea and to establish the differentiation strategies of breast cancer treatment.
As molecular analysis advances, many tests that predict a poor prognosis for breast cancer have been widely developed and used. Multigene assays that use tumor RNA, such as Oncotype DX [11] and MammaPrint [12], are helping to predict the risk of recurrence and metastasis, along with the benefit of adjuvant chemotherapy, in hormone receptor (HR)–positive breast cancer. In addition, molecular analysis using tumor DNA has been used to predict prognosis by identifying molecular alterations associated with poor prognosis, including single nucleotide variants (SNVs), small nucleotide insertions and deletions (Indels), copy number alterations (CNAs), and gene rearrangement. Genetic alterations in various genes are associated with poor prognosis, such as PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha) [13], ERBB2 [14], ESR1 (estrogen receptor 1) [15], and NF1 (neurofibromin 1) [16] and the genetic alterations have been widely analyzed by Sanger sequencing or real-time polymerase chain reaction. Next-generation sequencing (NGS) is an ideal tool for detecting molecular alterations associated with poor outcomes. It could help clinicians evaluate comprehensive molecular profiles of breast cancer and establish appropriate treatment plans. NGS also can provide numerous data about molecular alterations associated with target therapy.
The U.S. Food and Drug Administration (FDA) approved alpelisib for the treatment of patients with HR-positive/HER2-negative metastatic breast cancer and a PIK3CA mutation and olaparib for the treatment of patients with HR-positive/HER2-negative metastatic breast cancer with a germline BRCA mutation. Pembrolizumab was approved by the FDA to treat patients with microsatellite instability–high (MSI-H) solid tumors or solid tumors with a high tumor mutation burden (TMB). When treating patients with solid tumors that harbor NTRK gene fusion, larotrectinib and entrectinib can be administered. When the amount of information that can be provided is considered, an NGS study is cost-effective and has a relatively fast turnaround time because it can identify multiple target biomarkers at once. However, some researchers have suggested skeptical opinions about the need for NGS studies in daily practice, especially in breast cancer [17]. Therefore, it is crucial to accumulate real-world data of NGS studies and evaluate the effectiveness NGS in real-world clinical setting. In this study, we analyze real-world data about the prevalence of various genes used to prescribe targeted therapy for breast cancer and investigate how much target therapy has actually been applied.
Materials and Methods1. Patient selectionWe retrospectively collected and analyzed the histopathologic features and genetic profiles of 180 patients who were treated for breast cancer at Samsung Medical Center between 2010 and 2021 and underwent NGS studies of tissues collected during biopsy or surgery. Each patient’s initial clinical stage and pathologic stage were determined according to the criteria of the American Joint Committee on Cancer staging 8th edition [18] based on results from computed tomography (CT), magnetic resonance imaging, and positron emission tomography CT.
2. Histopathologic analysis and immunohistochemistryWe used formalin-fixed, paraffin-embedded (FFPE) tissue for histopathologic analysis and immunohistochemical (IHC) staining. All of the FFPE tissues were stained with hematoxylin and eosin. The detailed histopathologic features were reviewed by three breast pathologists (H.L., Y.A.C., and E.Y.C.). IHC staining was performed on 4-μm sections of FFPE tissue using a Leica BOND-III system (Leica Biosystems, Wetzlar, Germany) to stain for estrogen receptor (ER; 6F11, 1:300, Novocastra, Newcastle upon Tyne, UK), progesterone receptor (clone 16, 1:1,200, Novocastra), p53 (DO7, 1:1,200, Leica Biosystems), Ki-67, (MIB1, 1:200, Dako, Carpinteria, CA), and cytokeratin 5/6 (D5/16B4, 1:100, Dako) and the Ventana BanchMark XT platform (Ventana Medical Systems Inc., Tucson, AZ) to stain for HER2 (4B5, Ventana Medical Systems Inc.) according to the manufacturers’ protocols.
3. Next-generation sequencingTumor DNA was extracted from 5-μm FFPE sections using a QIAamp DSP DNA FFPE tissue kit (Qiagen, Hilden, Germany), and the extracted DNA was quantified using a QUBIT dsDNA BR assay kit (Thermo Fisher Scientific, Waltham, MA). For target capture, we used a SureSelect XT automation reagent kit (Agilent, Santa Clara, CA) according to the manufacturer’s protocol. A paired-end sequencing library was prepared using a barcode. To validate the size and quality of the genomic DNA, we used Genomic DNA Analysis ScreenTape and Genomic DNA Reagent with an Agilent 4200 tape station (Agilent). Libraries were sequenced using a TG NextSeq 500/550 high output kit v2 (Illumina, Inc., San Diego, CA) and a TG NextSeq 500/550 mid output kit v2 (Illumina, Inc.).
Seventy cases collected for NGS study from March 2018 to October 2020 were analyzed using a customized targeted NGS panel called CancerSCAN, which targets 375 genes associated with solid tumors. The list of target genes is provided in S1 Table. The other 110 cases collected for NGS study from November 2020 to July 2022 were analyzed using a hybrid capture–based TruSight Oncology (TSO) 500 DNA/RNA NextSeq kit (Illumina, Inc.). The list of 523 target genes for this kit is provided in S2 Table. Both CancerSCAN and TSO500 were performed by the same technician and the results were interpreted by the same pathologists (H.L. and E.Y.C.).
TMB was defined as the number of somatic mutations per megabase (Mb). A TMB less than 10 was defined as low-TMB, and a TMB of 10 or more was classified as high-TMB. To calculate TMB appropriately, all synonymous SNVs, Indels, CNAs, and fusion variants were excluded. Detailed information about the calculation is provided in S3 Table.
In cases tested with the TSO500, MSI status was evaluated according to the manufacturer’s instructions using 130 homopolymer microsatellite loci. When the percentage of unstable sites was 20 or more, the case was classified as MSI-H.
4. Statistical analysisAll statistical analyses were performed using the IBM SPSS statistical software package, ver. 27 (IBM Corp., Armonk, NY). Pearson’s chi-square tests and Fisher’s exact tests were performed to compare categorical variables, and paired t tests were performed to compare continuous variables. The Shapiro-Wilk test, Mann-Whitney U test, and ANOVA were performed to compare non-normally distributed variables. Survival data were analyzed using the Kaplan-Meier method and compared with the log-rank test. A p-value lower than 0.05 was considered statistically significant.
Results1. Clinicopathological patient characteristicsThe clinicopathological characteristics of the patients and information about the samples are summarized in Table 1. Only four patients (2.2%) were male, and the remaining 176 patients were female. The mean age of the patients was 41.46 years, and 117 patients (65.0%) were premenopause. Thirty-four patients showed local recurrence or distant metastasis after adjuvant chemotherapy, and 136 patients showed disease progression during neoadjuvant chemotherapy (NAC) or palliative therapy. Among the remaining 10 patients, nine patients underwent NAC and showed a partial response, and one patient did not receive NAC. None of those 10 patients showed any evidence of recurrence or distant metastasis during follow-up. The mean follow-up period was 64.02 months (range, 6.20 to 289.87 months). During follow-up, 77 patients died of the disease, and 81 patients lived with carcinoma. Only 22 patients had no evidence of disease. The most common histologic diagnosis was invasive breast cancer of no special type (159/180, 88.3%), followed by metaplastic carcinoma (14/180, 7.8%) and invasive lobular carcinoma (4/180, 2.2%). After dividing cases into subtypes based on the IHC results, 96 cases (53.3%) were triple-negative breast cancer (TNBC), and 72 cases (40.0%) were HR-positive, HER2-negative breast cancer. Within our study population, 62 patients (34.4%) underwent NGS during palliative chemotherapy, 58 after NAC, 34 after surgery and adjuvant chemotherapy, and 26 at the time of diagnosis.
Among the study samples, 95 (52.8%) were obtained from the primary breast cancer during surgery (66, 69.5%) or biopsy (39, 30.5%). The other 85 samples (47.2%) were obtained from metastatic breast cancer. Among the cases obtained from metastatic breast cancers, 60 samples (70.6%) were obtained by biopsy, and 25 cases (29.4%) were obtained during surgery. The most common metastatic site was the liver (27/85, 31.8%), followed by the lymph node (14/85, 16.5%), lung (12/85, 14.1%), pleura (12/85, 14.1%), and brain (6/85, 7.1%).
2. Molecular alterationsThe landscape of genetic alterations is summarized and visualized in Fig. 1. The most frequently detected SNV was the TP53 mutation (123/180, 68.3%), followed by the PIK3CA mutation (51/180, 28.3%). Among the CNAs, MYC amplification (36/180, 20.0%) was found most frequently, followed by CCND1 amplification (22/180, 12.2%). The detailed prevalence of molecular alterations in HR-positive breast cancer and TNBC are summarized in Tables 2 and 3. We compared the prevalence of molecular alterations in our study with those from The Cancer Genome Atlas (TCGA) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), which were accessed via cBioPortal [19-23]. In HR-positive breast cancer, the frequency of SNVs of ESR1 was statistically higher compared to TCGA and that of TP53 was statistically higher compared to both TCGA and METABRIC (Benjamini-Hochberg adjusted p-value < 0.05). The frequency of CNAs in HR-positive breast cancer and the frequency of SNVs and CNAs in TNBC was not significantly different.
3. PIK3CA mutationA PIK3CA SNV was detected in 28.3% (51/180) of our patients, and the amplification of PIK3CA was detected in one patient. Detailed information about the PIK3CA SNVs is summarized in Table 4. The most frequent PIK3CA mutation was H1047R (43.1%, 22/51), followed by E545K (17.6%, 9/51), N345K (13.7%, 7/51), and E542K (9.8%, 5/51). Other mutations were found in one patient. In patients with HR-positive breast cancer, 31/72 (43.1%) had a PIK3CA mutation, and 11/96 (11.5%) patients with TNBC had a PIK3CA mutation. Nine patients with HER2-positive breast cancer (9/12, 75%) had a PIK3CA mutation. The detailed distribution of PIK3CA mutations in HR-positive breast cancer, HER2-positive breast cancer, and TNBC is visualized in Fig. 2. The proportion of PIK3CA mutations included in the therascreen panel was 72.5%. Among PIK3CA mutations not included in the therascreen panel, N345K was the most frequently detected (50.0%) (Fig. 3). After adjustment based on the IHC subtypes, 34.7% (25/72) of patients with HR-positive, HER2-negative breast cancer were expected to benefit from a phosphoinositide 3-kinase (PI3K) pathway inhibitor.
4. BRCA1/2 mutationA BRCA1 mutation was found in six patients (3.3%), and a BRCA2 mutation was found in 13 patients (7.2%). To confirm the germline mutation, matched normal blood samples were collected from 84 patients, and BRCA1/BRCA2 sequencing was performed. In those 84 patients, a BRCA1 germline mutation was found in four patients, and a BRCA2 germline mutation was found in five patients, all of which were also detected by NGS. The four patients with a BRCA2 mutation that was detected only by NGS were confirmed to have somatic mutations. The remaining two of six patients with a BRCA1 mutation detected in NGS and four of 13 patients with a BRCA2 mutation detected in NGS did not receive BRCA1/BRCA2 sequencing using matched blood samples.
5. ESR1 mutation
ESR1 mutations were detected in 11 patients (6.1%), of whom 10 (5.5%) had HR-positive, HER2-negative breast cancer and the other had TNBC. Detailed information about patients with an ESR1 mutation is summarized in Table 5. Two patients with HR-positive, HER2-negative breast cancer and one patient with TNBC had not previously been treated with endocrine therapy. One patient had two ESR1 mutations, E380Q/Y537D. The details of the ESR1 mutations were as follows: SNVs in eight cases; D538G in five cases, E380Q/Y537D in one case, L536H in one case, L536_Y537insH in one case, fusion in three cases, ESR1:CCDC170 fusion in 2 cases, and ESR1:TAB2 fusion in one case (S4 Table).
6. Application of precision medicines based on NGS resultsThirteen patients (13/180, 7.2%) received target therapy based on the results of their NGS studies. That information is detailed in Table 6. Four patients had a BRCA1 or BRCA2 germline mutation, and they received PARP inhibitor therapy (olaparib or talazoparib). In patients with BRCA mutation, three patients received treatment as part of a clinical trial, and another patient was administered target therapy through expanded access of emergency investigational new drug (e-IND). The other nine patients had PIK3CA mutations and received PI3K pathway inhibitor therapy (alpelisib). Among the nine patients with a PIK3CA mutation, six patients could access alpelisib treatment via e-IND, and the other three patients required an additional molecular analysis (QIAGEN’s therascreen PIK3CA RGQ, a companion diagnostic tool for alpelisib).
7. Tumor mutation burdenThe mean TMB was 7.37/Mb (range, 0 to 68.7), and the median TMB was 6.3/Mb. TMB did not differ significantly according to age (≤ 50 years vs. > 50 years, 7.03/Mb vs. 8.29/Mb, p=0.426), menopause (premenopause vs. postmenopause, 6.93/Mb vs. 8.29/Mb, p=0.329), whether the samples were obtained from a metastasis (primary vs. metastasis, 8.02/Mb vs. 6.64/Mb, p=0.168), whether the samples were obtained after treatment (initial vs. post-treatment, 6.74/Mb vs. 7.48/Mb, p=0.506), NGS timing (initial vs. after NAC vs. at recurrence vs. during palliative chemotherapy, 6.74/Mb vs. 7.05/Mb, vs. 6.38/Mb vs. 8.48/Mb, p=0.448) or subtype (HR-positive, HER2-negative vs. HER2-positive vs. TNBC, 7.73/Mb vs. 9.98/Mb vs. 6.78/Mb, p=0.270). Eleven cases had high TMB (6.1 %). The mean TMB of the high-TMB cases was 26.54 (range, 10.90 to 68.70; median, 22.00). None of the cases that underwent the TSO500 panel were classified as MSI-H or MSI-Low.
DiscussionNGS is an ideal method for comprehensive molecular profiling of solid tumors. Given the amount of information provided, NGS is relatively cost-effective and has a fast turnaround time. It has opened the era of precision medicine by detecting molecular alterations in a wide genomic region that can predict a patient’s prognosis or treatment effect and guide clinicians in administering an appropriate target therapy. In this study, we collected data from our institute to determine the effectiveness of NGS and present real-world data about precision medicine based on NGS results.
The drug-targetable biomarker most commonly found in our study was PIK3CA mutations, which were found in 43.1% (31/72) of HR-positive, HER2-negative breast cancer patients and 11.5% (11/96) of TNBC patients. The SOLAR-1 clinical trial showed that alpelisib plus fulvestrant significantly prolonged progression-free survival (PFS) in HR-positive, HER2-negative breast cancer patients with a PIK3CA mutation [25]. The SOLAR-1 trial included patients with PIK3CA hotspot mutations in the C2, helical, and kinase domains (corresponding to exons 7, 9, and 20, respectively) as found using QIAGEN’s therascreen PIK3CA RGQ kit, and the FDA approved that PCR kit as a companion diagnostic device for alpelisib. In this study, 72.5% (37/51) of cases with a PIK3CA mutation had PIK3CA SNVs included in that kit, and the other 27.5% (14/51) had mutations not included in the kit [24]. The N345K and Q546K mutations showed sensitivity to PI3K pathway inhibitors in preclinical models [26]. Therefore, additional studies are needed to determine whether breast cancer patients with other rare variants that are not included in the therascreen PIK3CA RGQ PCR kit show therapeutic benefit from receiving PI3K pathway inhibitors. A sufficient investigation should be conducted to allow more patients with advanced HR-positive, HER2-negative breast cancer and PIK3CA mutations to benefit from targeted therapy.
Another important drug-targetable biomarker is BRCA1/BRCA2 mutation. BRCA1 and BRCA2 are genes associated with the risk of ovarian cancer and breast cancer. Excluding cases in which germline BRCA1/BRCA2 mutations could not be confirmed, a BRCA1 germline mutation was found in 4.8% (4/84) of the patients in this study, and a BRCA2 germline mutation was found in 6.0% (5/84) of patients. No somatic mutations of BRCA1 were found, but somatic mutations of BRCA2 were found in 4.8% (4/84) of patients.
Olaparib has been approved by the FDA for use as adjuvant chemotherapy in patients with germline BRCA mutations in high-risk, HER2-negative breast cancer or as palliative chemotherapy in patients with germline BRCA1/2 mutations in metastatic breast cancer. The National Comprehensive Cancer Network guideline also suggests that olaparib can be useful in some patients with somatic BRCA mutations in metastatic breast cancer. In addition to anti-cancer treatment, it has been reported that prophylactic mastectomy and salpingo-oophorectomy reduce the lifetime cancer risk and increase survival times in patients with a germline BRCA1/2 mutation [27]. Because a BRCA1/2 mutation is a dominant inheritance, familial history should be evaluated, and genetic consultations should be used to determine the lifetime cancer risk of family members. In breast cancer patients, BRCA1/2 mutation evaluations must be performed through germline BRCA1/2 sequencing or NGS.
In addition to drug-targetable biomarkers, NGS can provide information about molecular alterations that can predict therapy resistance. ER with a hotspot mutation in the ligandbinding domain (LBD) of ESR1 undergoes a conformational alteration to maintain an agonistic active conformation similar to estrogen-bound wild-type ER [28]. ER with genomic rearrangement of ESR1 thus creates a chimeric protein containing a proximal promoter domain without an LBD, resulting in the production of a constitutive promoter and ligand-independent tumor growth [28]. Therefore, an ESR1 mutation in HR-positive breast cancer can predict resistance to endocrine therapy [15,29,30]. ESR1 mutations are acquired after the administration of endocrine therapy, especially aromatase inhibitors, but some primary breast cancer without a history of endocrine therapy can also harbor an ESR1 mutation [28]. In our study population, 3 of 11 cases with ESR1 mutations had not received endocrine therapy before tissue sampling, suggesting that ESR1 mutations can be found in primary breast cancer without prior exposure to aromatase inhibitors.
An ESR1 mutation can be used as a predictive biomarker, as well as a drug-targetable biomarker. Recently, the EMERALD trial reported that an oral selective ER degrader, elacestrant, showed significant PFS improvement in patients with advanced/metastatic HR-positive/HER2-negative ESR1-mutant breast cancer who had progressed during prior treatment with endocrine therapy or CDK4/6 inhibitor therapy [31]. Therefore, an evaluation for an ESR1 mutation is crucial not only to predict endocrine therapy resistance, but also to enable the administration of appropriate targeted therapy.
TMB is another type of important information that NGS can provide. Many studies have reported that high TMB is associated with the production of many neoantigens and increases T cell reactivity, suggesting that it might be closely related to an improved immune checkpoint inhibitor (ICI) response [32,33]. The clinical benefit of ICI administration has been reported in patients with breast cancer, especially TNBC [6,7], and predictive biomarkers of an ICI response such as programmed death-ligand 1 are in the limelight as an essential evaluation in real clinical settings. However, TMB-high breast cancer was reported to be very rare [34], and our study confirms that only 6.1% of tumors had TMB ≥ 10. No MSI-H tumors were found.
For an accurate TMB evaluation, it is necessary to exclude germline variants by using matched normal tissues or blood. However, it is difficult to analyze both tumor tissue and matched normal tissues together in a real-world clinical setting because of financial and time constraints. TMB might be overestimated in a tumor-only NGS study unless germline variants or FFPE-induced deamination artefacts are properly excluded. Therefore, SNVs or Indels with a high probability of being germline variants should be carefully excluded using population frequencies in a well-known germline database [35]. Further studies should be conducted to establish guidelines for classifying variants as germline or somatic by setting appropriate cut-off values for population frequency. In addition, appropriate standards for total depth and variable frequency alleles should be established to exclude spurious variants generated by artefacts.
Recently, various groups have expressed skepticism about the need for NGS studies in daily practice. It is difficult to apply NGS to all patients because it is more time-consuming and expensive than other screening tools. For example, PIK3CA mutations in breast cancer, which are drug-targetable biomarkers, can be detected by real-time PCR; HER2 can be evaluated by IHC and in situ hybridization, and germline variants of BRCA1/2 can be evaluated by blood sequencing [17]. Our results indicate that only 7.2% of patients benefited from their NGS study, and were not significantly different from previous studies in South Korea [36]. Also 33% (3/9) of patients with a PIK3CA mutation had to undergo an additional companion diagnostic test. However, NGS studies can provide many predictive biomarkers can predict aggressive behavior by breast cancer, such as PIK3CA [13] and ERBB2 [14] mutations, or resistance to endocrine therapy, such as ESR1 [15] and NF1 [16] mutations. In addition, NGS examinations can provide new therapeutic opportunities to patients with metastatic or advanced breast cancer by detecting genomic alterations that match the requirements of clinical trials testing targeted therapies. Our study revealed that among 13 patients who benefited from precision medicine based on NGS study, 76.9 % (10/13) of patients received target therapy through clinical trial or expanded accecss of e-IND. These results demonstrated the effectiveness of NGS studies in enabling clinicians to provide target therapies as part of a clinical trials or e-IND to patients, especially advanced breast cancer patients who are refractory to most of standardized treatments and have extremely limited available treatment options.
In conclusion, our study has revealed that only 7.2% of breast cancer patients who underwent NGS in a real-world clinical setting benefited from the test. Given financial and time constraints, the application of NGS to all patients with breast cancer in daily practice might be excessive. However, it is possible to forecast prognoses of breast cancer by discovering predictive biomarkers that suggest aggressive behavior and resistance to endocrine or target therapy and by detecting potential drug-targetable biomarkers that can offer patients the chance to participate in a clinical trial. Therefore, the value of NGS in advanced or metastatic breast cancer could be increased through further studies.
Electronic Supplementary MaterialSupplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
NotesEthical Statement Our study was approved by the Institutional Review Board of Samsung Medical Center (IRB No. 2022-09-038). Informed written consent from patients was waived by the Institutional Review Board of Samsung Medical Center because of the retrospective study design. Fig. 1.Schematic demonstration of clinicopathological characteristics and genetic alterations. ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; TMB, tumor mutation burden; TNBC, triple-negative breast cancer. 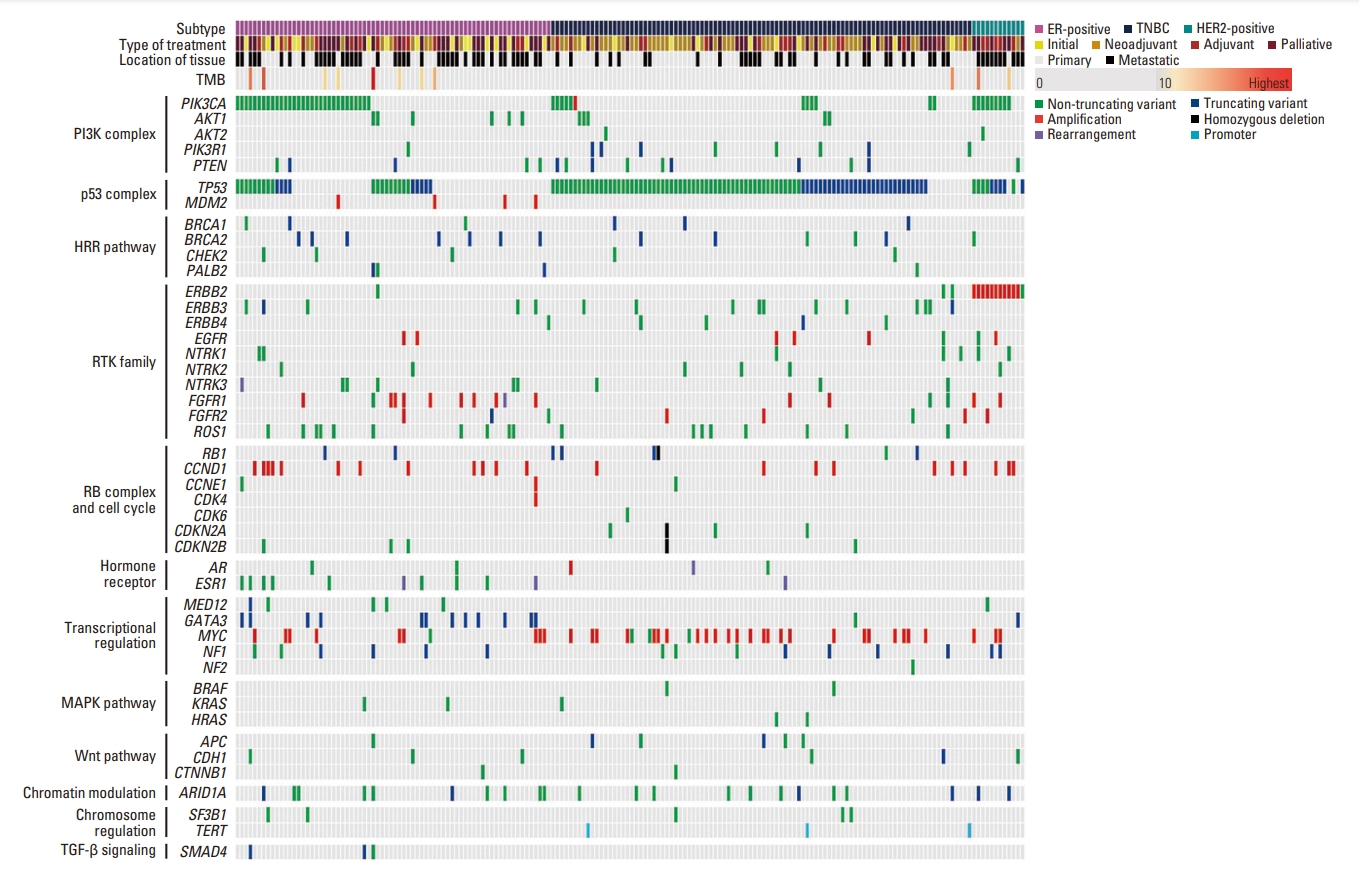
Fig. 2.The distribution of PIK3CA single nucleotide variants (SNVs) based on immunohistochemical subtypes. (A) Hormone receptor–positive, human epidermal growth factor receptor 2 (HER2)–negative breast cancer. (B) HER2-positive breast cancer. (C) Triple-negative breast cancer. The most frequent SNV in all subtypes was H1047R. 
Fig. 3.The distribution of PIK3CA single nucleotide variants (SNVs) based on the therascreen panel. (A) The prevalence of PIK3CA SNVs included in the therascreen panel. (B) The distribution of PIK3CA SNVs not included in the therascreen panel. 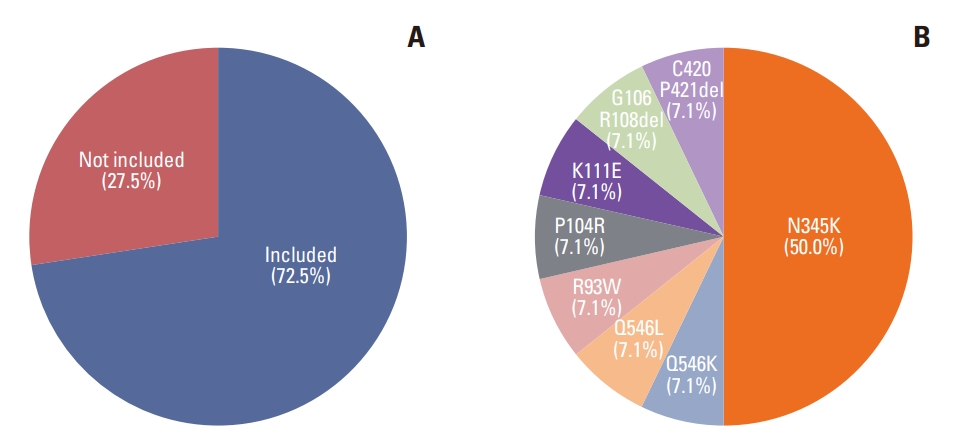
Table 1.Clinicopathologic features of 180 cases Table 2.Frequency of SNVs and Indels in breast cancers reported in literature
Values are presented as number of cases with SNV and Indel (%) unless otherwise noted. Indel, insertion and deletion; SNV, single nucleotide variant; TCGA, The Cancer Genome Atlas. a) Public data are adopted from cBioPortal (https://www.cbioportal.org/). Table 3.Frequency of amplification in breast cancers reported in literature
Values are presented as number of cases with amplification (%) unless otherwise noted. TCGA, The Cancer Genome Atlas. a) Public data are adopted from cBioPortal (https://www.cbioportal.org/). Table 4.The detailed information of PIK3CA mutation
a) The clinical significance of mutation was adopted from Clinvar (https://www.ncbi.nlm.nih.gov/clinvar/) and OncoKB (https://www.oncokb.org) [24]. Table 5.Clinical characteristics of breast cancer patients with ESR1 alterations Table 6.Treatment of patients who received target therapy based on precision medicine
References1. Choi JE, Kim Z, Park CS, Park EH, Lee SB, Lee SK, et al. Breast cancer statistics in Korea, 2019. J Breast Cancer. 2023;26:207–20.
2. Hudis CA. Trastuzumab: mechanism of action and use in clinical practice. N Engl J Med. 2007;357:39–51.
3. Modi S, Saura C, Yamashita T, Park YH, Kim SB, Tamura K, et al. Trastuzumab deruxtecan in previously treated HER2-positive breast cancer. N Engl J Med. 2020;382:610–21.
5. Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16:25–35.
6. Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379:2108–21.
7. Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im SA, Yusof MM, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. 2020;396:1817–28.
8. Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A, et al. Breast cancer statistics, 2022. CA Cancer J Clin. 2022;72:524–41.
9. Shin HC, Lee HB, Yoo TK, Lee ES, Kim RN, Park B, et al. Detection of germline mutations in breast cancer patients with clinical features of hereditary cancer syndrome using a multi-gene panel test. Cancer Res Treat. 2020;52:697–713.
10. Han SA, Park SK, Ahn SH, Lee MH, Noh DY, Kim LS, et al. The Korean Hereditary Breast Cancer (KOHBRA) study: protocols and interim report. Clin Oncol (R Coll Radiol). 2011;23:434–41.
11. Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379:111–21.
12. Piccart M, van ‘t Veer LJ, Poncet C, Lopes Cardozo JM, Delaloge S, Pierga JY, et al. 70-gene signature as an aid for treatment decisions in early breast cancer: updated results of the phase 3 randomised MINDACT trial with an exploratory analysis by age. Lancet Oncol. 2021;22:476–88.
13. Li SY, Rong M, Grieu F, Iacopetta B. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat. 2006;96:91–5.
14. Wang T, Xu Y, Sheng S, Yuan H, Ouyang T, Li J, et al. HER2 somatic mutations are associated with poor survival in HER2-negative breast cancers. Cancer Sci. 2017;108:671–7.
15. Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45:1439–45.
16. Zheng ZY, Anurag M, Lei JT, Cao J, Singh P, Peng J, et al. Neurofibromin is an estrogen receptor-alpha transcriptional co-repressor in breast cancer. Cancer Cell. 2020;37:387–402.
17. Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31:1491–505.
18. Amin MB, Edge SB, Greene FL, Byrd DR, Brookland RK, Washington MK, et al. AJCC cancer staging manual. 8th ed. New York, NY: Springer; 2017. p. 589–628.
19. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479.
20. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52.
21. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
22. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
23. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
24. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017:PO.17.00011.
25. Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380:1929–40.
26. Dogruluk T, Tsang YH, Espitia M, Chen F, Chen T, Chong Z, et al. Identification of variant-specific functions of PIK3CA by rapid phenotyping of rare mutations. Cancer Res. 2015;75:5341–54.
27. Ludwig KK, Neuner J, Butler A, Geurts JL, Kong AL. Risk reduction and survival benefit of prophylactic surgery in BRCA mutation carriers, a systematic review. Am J Surg. 2016;212:660–9.
28. Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations: a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12:573–83.
29. Jeselsohn R, Yelensky R, Buchwalter G, Frampton G, Meric-Bernstam F, Gonzalez-Angulo AM, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20:1757–67.
30. Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–51.
31. Bidard FC, Kaklamani VG, Neven P, Streich G, Montero AJ, Forget F, et al. Elacestrant (oral selective estrogen receptor degrader) versus standard endocrine therapy for estrogen receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: results from the randomized phase III EMERALD trial. J Clin Oncol. 2022;40:3246–56.
32. Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21:1353–65.
33. Winer EP, Lipatov O, Im SA, Goncalves A, Munoz-Couselo E, Lee KS, et al. Association of tumor mutational burden (TMB) and clinical outcomes with pembrolizumab (pembro) versus chemotherapy (chemo) in patients with metastatic triple-negative breast cancer (mTNBC) from KEYNOTE-119. J Clin Oncol. 2020;38(15 Suppl):1013.
34. Barroso-Sousa R, Jain E, Cohen O, Kim D, Buendia-Buendia J, Winer E, et al. Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann Oncol. 2020;31:387–94.
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||