AbstractPurposeRecently, we developed allele-discriminating priming system (ADPS) technology. This method increases the sensitivity of conventional quantitative polymerase chain reaction up to 100 folds, with limit of detection, 0.01%, with reinforced specificity. This prospective study aimed to develop and validate the accuracy of ADPS epidermal growth factor receptor (EGFR) Mutation Test Kit using clinical specimens.
Materials and MethodsIn total 189 formalin-fixed paraffin-embedded tumor tissues resected from patients with non–small cell lung cancer were used to perform a comparative evaluation of the ADPS EGFR Mutation Test Kit versus the cobas EGFR Mutation Test v2, which is the current gold standard. When the two methods had inconsistent results, next-generation sequencing–based CancerSCAN was utilized as a referee.
ResultsThe overall agreement of the two methods was 97.4% (93.9%-99.1%); the positive percent agreement, 95.0% (88.7%-98.4%); and the negative percent agreement, 100.0% (95.9%-100.0%). EGFR mutations were detected at a frequency of 50.3% using the ADPS EGFR Mutation Test Kit and 52.9% using the cobas EGFR Mutation Test v2. There were 10 discrepant mutation calls between the two methods. CancerSCAN reproduced eight ADPS results. In two cases, mutant allele fraction was ultra-low at 0.02% and 0.06%, which are significantly below the limit of detection of the cobas assay and CancerSCAN. Based on the EGFR genotyping by ADPS, the treatment options could be switched in five patients.
IntroductionDue to progress in cancer genetics and genomics, cancer management has entered the era of personalized-precision medicine [1,2]. Inaccurate genotyping may lead to a faulty or suboptimal treatment. Therefore, there is growing need for platforms with a high diagnostic [3,4]. Advancements in genotyping technology resulted in the development of multiplex epidermal growth factor receptor (EGFR) mutation analysis platforms with high specificities and sensitivities; 1% limit of detection (LoD) for in cobas EGFR Mutation Test v2 [5], 0.81% in therascreen EGFR RGQ PCR Kit [6], 0.1% in droplet digital polymerase chain reaction (PCR) [7], 0.5% in next-generation sequencing–based CancerSCAN [8,9], and 0.01% in SuperSelective PCR [10,11]. Recently, we developed allele-discriminating priming system (ADPS), a novel method that can accurately detect rare mutations based on genetically modified Taq DNA polymerase that improves the specificity between primers and templates during quantitative polymerase chain reaction (qPCR) amplification [12]. To translate the technological renovations into clinical application, the ADPS EGFR multiplex assay was developed, and comparative analyses of clinical samples were performed to evaluate its applicability at points of care.
The discovery of activating mutations in the tyrosine kinase domain of EGFR caused a paradigm shift in understanding the molecular biology of non–small cell lung cancer (NSCLC) and in the use subsequent treatment strategies [13-16]. Approximately 45% of EGFR mutations are exon 19 deletions, and 40%-45% are exon 21 L858R substitutions [17-19]. All the activating EGFR somatic mutations involve the adenosine triphosphate binding area in the receptor tyrosine kinase domain, to which tyrosine kinase inhibitor (TKI) is binding [20]. Randomized trials have shown that in terms of efficacy, TKIs are superior to platinum-based cytotoxic chemotherapy, which was the standard of care for patients with treatment-naïve EGFR-mutated NSCLC [21-23]. Thus, an accurate diagnosis of EGFR mutation is a key for successful therapies in lung cancer.
However, patients frequently develop resistance to drugs and experience disease progression at a median of 9-13 months after EGFR-TKI treatment [24]. Among the acquired resistance mechanisms, T790M mutation is a well-evaluated mechanism of acquired resistance. Osimertinib, a third-generation EGFR-TKI targeting both conventional EGFR-TKI–sensitizing mutations and resistant T790M mutations, is significantly effective when used as the first-line therapy with a median overall survival of 38.6 months [16]. Therefore, there is an increasing demand to accurately identify rare T790M mutation. Moreover, in addition to exon 19 deletions and exon 21 L858R missense, several uncommon yet activating EGFR mutations, including S768I, L861Q, and exon 20 insertions account for 10%-20% of all EGFR mutations [25]. Interestingly, as these mutations demonstrated different responses to first, second, and third-generation EGFR-TKIs [15,21,26], an accurate EGFR genotyping is important in therapeutic decision-making for NSCLC.
This prospective blinded study aimed to develop ADPS EGFR Mutation Test Kit (ADPS assay) using the ADPS technology, which is designed to detect the aforementioned actionable EGFR mutation in patients with suspicious lung cancer at the clinic. Further, to evaluate its accuracy, the cobas EGFR Mutation Test v2 (cobas assay) was used as a reference assay platform and next-generation sequencing (NGS)–based multiplexed cancer mutation assay, named CancerSCAN [8,9], as an additional assay platform.
Materials and Methods1. Primer design for the multiplex EGFR assayThe 3′ terminal base of each allele-specific primer was designed according to its corresponding mutation or allele sequence. In case of indels, 2-4 mismatches were designed at the 3’ end to distinguish the primer sequences according to the sequence difference before and after insertion or deletion. The allele-specific primer was designed at a melting temperature (< 60°C) to increase selectivity due to mismatch at the 3′ end. However, the primer used on the other side of the allele-specific primer was designed at a similar temperature or slightly higher than 60°C for sufficient binding efficiency. The melting temperature of oligonucleotides was calculated using OligoAnalyzer (Integrated DNA Technologies, Coralville, IA). Oligonucleotides were synthesized either by Integrated DNA Technologies or Biosearch Technologies (Hoddesdon, UK). Optimal primers and probes were selected using the following criteria: First, they should not suppress mutant-specific PCR efficiency, can balance longer primer length and low melting temperature at approximately 56°C-58°C, which is lower than standard 60°C by 2°C-4°C, and have no dimer formed. Second, they should not allow non-specific amplification of wild-type (WT) template, with significant DCt between mutant- and WT templates, have a primer length that minimizes non-specific amplification (lower than the annealing temperature of PCR), and does not allowing false priming at the 3′ ends with proper GC-contents to prevent extension of short fragments. The detectable EGFR mutations of ADPS EGFR Mutation Test Kit are listed in S1 Table.
2. qPCR template preparationDNA fragments were synthesized to encompass each mutation and WT sequences (Bio Basic, Markham, Canada), and cloned into pBluescript II SK(+) plasmid. The synthesized sequence was confirmed via Sanger sequencing. Each plasmid template was linearized with a restriction enzyme at locations outside the insert and diluted to 109 copies/μL, via the calculation of molecular weights. To dilute the plasmid, TE buffer (AM9849, Invitrogen, Waltham, MA) containing 2 ng/μL of sheared salmon sperm DNA (AM9680, Invitrogen) was used. To validate the detection sensitivity in formalin-fixed paraffin-embedded (FFPE) specimen, FFPE reference standard were purchased and used as reference materials (EGFR S768I Reference Standard, EGFR L861Q Reference Standard, EGFR V769_D770insASV Reference Standard and EGFR Gene-Specific Multiplex Reference Standard for Ex19Del, T790M, G719S and L858R [HD707, HD133, HD706, and HD300 respectively, Horizon Discovery, Cambridge, UK]). Genomic DNA of the FFPE reference standard was extracted using the QIAamp DNA DSP FFPE Tissue Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Plasmid or genomic DNA concentrations were evaluated using the Epoch microplate spectrophotometer (BioTek Instruments, Winooski, VT), and genomic DNA copy numbers were calculated, assuming that one copy of the haploid genome is 3.3 pg. To dilute the genomic DNA, TE buffer (AM9849, Invitrogen) was used.
3. qPCR reaction conditionIn 1× ADPS reaction buffer, qPCR was performed with 0.25 mM of dNTP, 200 nM of forward primer, 200 nM of reverse primer, 400 nM of dual-labeled fluorescent probe, 2 μL of DNA template of desired copy number, and 0.5 U of ADPS Smart DNA polymerase (GENECAST, Seoul, Korea), with a total volume of 20 μL. The reactions were performed in the 7500 Fast Real-Time PCR System (Applied Biosystems, Waltham, MA), with an initial denaturation for 5 minutes at 95°C followed by thermal cycles: denaturation at 95°C for 10 seconds, annealing at 60°C for 30 seconds (fluorescence acquisition at this step), and elongation at 72°C for 10 seconds. All qPCR data were analyzed in 7500 software (Applied Biosystems).
4. LoD determinationTo evaluate the detection sensitivity of the ADPS assay, the following FFPE reference standard materials were used (Horizon Discovery): EGFR S768I (HD707), EGFR L861Q (HD133), EGFR Ex20Ins (c.2300_2308dup, HD706), and other mutations such as EGFR Ex19Del (c.2235_2239del15), T790M, G719S and L858R (HD300). To determine the LoD of the ADPS assay using the FFPE specimens, eight replicates were run three times using reagents from different lots each time (total: 24 replicates per specimen). If positive calls were reproduced at least in 23 of 24 replicates (reproducibility: > 95%), the mutation calls were considered reliable, and the lowest concentration among the reliable calls were determined as LoD of ADPS assay. Near LoD, multiple tests were conducted to confirm reliability.
5. Clinical specimensFormalin-fixed paraffin-embedded tissue (FFPET) sections were obtained from patients with histologically confirmed NSCLC who underwent surgical resection at Samsung Medical Center (Table 1). To determine accurate EGFR genotypes with a sufficient amount of tissue samples, surgical FFPET specimens were used in this study. Pathologic diagnosis and staging were performed according to the 8th American Joint Committee on Cancer Staging Manual. All FFPET sections were evaluated by pathologist to confirm tumor histology. The current study was approved by the institutional review board of Samsung Medical Center. All patients provided written informed consent for the procurement of tumor specimens.
Patients with stage Ib-IIIa disease who require postoperative adjuvant chemotherapy were selected for clinical validation. The additional selection criteria were minimal smoking history and tumor locations in the periphery. In total 195 patients were enrolled blindly before surgery (no knowledge on the nature of the tumors [malignant vs. benign], including cell types). Six patients were excluded from the study. Among them, five patients had benign lesions, which were confirmed via intraoperative frozen biopsy using wedge-resected specimens. Further, one patient withdrew from the study. Based on clinical practice in Samsung Medical Center, FFPE samples were prepared from the tumor specimens. FFPET slides were prepared, and the tumor contents were determined by histological examination using hematoxylin and eosin staining at the pathology department of Samsung Medical Center (S2 Table). A sufficient amount of DNA was isolated from the FFPE samples, which were divided into three aliquots (one for the cobas assay, another for the ADPS assay, and the other for the NGS-based multiplex mutation assay if necessary). The whole assay procedure was performed blindly. That is, the service providers of the cobas assay and ADPS assays were blinded from each other, and from the identification of clinical samples.
6. EGFR mutation analysesTissue specimens were collected and prepared as FFPET samples. gDNA was extracted from FFPET using the QIAamp DNA FFPE Tissue Kit (Qiagen) for EGFR mutation analysis using the ADPS assay and cobas assay (Roche Molecular Systems, Pleasanton, CA). In total 50 ng of gDNA was diluted to 10 ng/μL or 2 ng/μL for the ADPS and cobas reactions. ADPS reactions were run in the 7500 Fast Real-Time PCR System (Applied Biosystems), and cobas reactions were run in the cobas 4800 system (Roche Molecular Systems, Pleasanton, CA). Both assays were performed according to the manufacturer’s protocols.
7. NGS-based CancerSCAN analysis of discrepant resultsNGS-based CancerSCAN analysis, originally designed at Samsung Medical Center in 2015 [8], was performed to analyze discrepant results between the ADPS assay and the cobas assay. CancerSCAN has processed more than 50,000 tests, and has acquired certification from the College of American Pathologists with a strict quality management system complying with the International Organization for Standardization (ISO) guidelines (ISO/TS 22692:2020 Health informatics - Quality control metrics for DNA sequencing). The CancerSCAN panel was designed to sequence 375 cancer-related genes. Further, 250 ng of genomic DNA was fragmented using the S220 Focused-ultrasonicator (Covaris, Woburn, MA), and target-capture was performed with the SureSelectXT Reagent Kit, HSQ (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s instruction.
After enrichment of exome libraries, they were sequenced in the HiSeq 2500 system (Illumina, San Diego, CA). Briefly, paired-end DNA sequencing libraries were prepared via gDNA shearing, end-repair, A-tailing, paired-end adaptor ligation and amplification. After hybridization of the libraries with bait sequences for 27 hours, the captured libraries were purified and amplified with an index barcode tag, and the quality and quantity of library were analyzed. The exome libraries were sequenced using the 100 bp paired-end mode of the TruSeq Rapid PE Cluster Kit and the TruSeq Rapid SBS Kit (Illumina). Sequence reads were mapped to the human genome (hg19) using the Burrows-Wheeler Aligner. Duplicated read was removed with Picard and SAMtools, and local alignment was optimized with the Genome Analysis Tool Kit (GATK). Variant calls for SNVs, small indels, copy number variations, and gene fusion in the targeted regions were analyzed.
Results1. Development of ADPS-based EGFR mutation assayThe ADPS technology is a type of allele-specific quantitative polymerase chain reaction, where the ADPS Smart DNA polymerase significantly improves allele-discrimination. In developing the EGFR mutation assay system, the cobas EGFR Mutation Test v2 (Roche Diagnostics, Basel, Switzerland) which is the standard for EGFR mutation detection worldwide, was used as a reference model. To detect several targeted EGFR mutations including SNV, insertion, and deletion, we designed allele-specific primers corresponding to the types and sequences of each mutation and confirmed the efficiency of the primer based on our previous study [12]. The ADPS assay and cobas assay comprise four and three multiplexes, respectively, that can detect 42 EGFR mutations, classified into seven mutation groups, including drug-response groups (such as Ex19Del, L858R, and T790M). Meanwhile, the cobas assay was designed and generated amplicons of 104-143 bp, the ADPS assay produced shorter amplicons of 61 to 104 bp which might amplify partially degraded gDNA from FFPE specimen more efficiently. To detect the seven mutation groups present in exons 18-21 of EGFR with different fluorescent signals, S768I, T790M, and Ex20Ins present in exon 20 of EGFR must be configured to react in different tubes. rs1050171 (A/G), a single nucleotide polymorphism (SNP), exists at an adjacent position to T790M, and the presence or absence of the SNP might affect the amplification. To prevent this phenomenon, probes corresponding to two types of sequences were designed and used. In case of mutation groups such as Ex19Del, Ex20Ins, G719X, and L858R, each mutation-specific allele-specific primer was designed with a common dual-labeled oligonucleotide probe, since there are two or more types of mutations (Fig. 1).
2. Analytical performance of ADPS-based EGFR mutation assayCell line gDNA containing mutated EGFR was used to confirm the validity of the constructed EGFR multiplex assay. The samples were diluted to 25.6%, 6.4%, 1.6%, 0.4%, and 0.1% mutant allele fraction (MAF) for each EGFR mutation, and the linearity between MAF and Ct was confirmed using the EGFR multiplex assay (Fig. 2). In the seven targets tested, linearity was validated in all sections from 25.6% to 0.1% (0.986 ≤ R2 ≤ 0.998). Hence, this assay had a high sensitivity and specificity.
To evaluate the analytical performance of the ADPS assay, reference standard materials in FFPE format were used (Horizon Discovery). The LoD, defined as the lowest concentration detected in > 95% of 24 replicates, were 0.01% for S768I, L861Q, and Ex20Ins, 0.02% for L858R, 0.05% for Ex19Del and T790M, and 0.2% for G719S (S3 Table) where 0.01% indicate three copies of EGFR-mutated DNA among 30,000 copies of total DNA. This result was consistent to the result using plasmid (S4 Fig.).
3. Performance of ADPS assay using clinical specimensThe accuracy of ADPS multiplex EGFR mutation assay was further investigated using clinical samples via a prospective blinded study. In total 189 patients were analyzed using the assays (Table 1). For comparative evaluations of various platforms with a plenty amount of samples, we selected stage Ib-IIIa resected tissues, and used FFPET samples prepared for the standard cobas assay. Tumor specimens analyzed using the ADPS assay and the cobas assay, generated highly congruent results: EGFR mutations were identified in 95 specimens (50.3%) using the ADPS assay, and 100 specimens (52.9%), using the cobas assay. L858R was the most prevalent mutation detected in both assays, and the combined frequencies of Ex19Del and L858R were 91.6% (ADPS assay) and 90.0% (cobas assay), respectively. Using the cobas assay as a reference model, the positive percent agreement was 95.0% (88.7%-98.4%), the negative percent agreement was 100.0% (95.9%-100.0%) and overall percent agreement was 97.4% (93.9%-99.1%). Cohen’s Kappa value was 0.95 (0.88-0.98), indicating considerably high agreement between the two assays (Table 2). The five discrepant samples listed in Table 2 were categorized based on the presence or absence of EGFR mutations in each specimen. For example, in the case of specimen CT-003, the ADPS assay detected only Ex19Del, while the cobas assay detected both Ex19Del and Ex20Ins. Since both assays detected an EGFR mutation, they were classified as a match. It should be noted that these differ from the 10 discrepancies in specific mutations that are discussed below.
4. Concordance and discrepancies between the ADPS assay and the cobas assayThere were 10 discrepant results between the ADPS assay and the cobas assay, despite an overall percent agreement of 97.4%. All 10 discrepancies were reproduced in repeated assays.
Next, to resolve the discrepancies in these 10 samples, the reserved third aliquots of DNA were used for CancerSCAN. Although CancerSCAN cannot be considered a definitive answer, it might be a highly qualified referee with a high reliability [8,9]. According to CancerSCAN results, the ten discrepant cases were grouped into three categories (Table 3). Both the ADPS assay and CancerSCAN did not detect any mutation in subset I (n=7/10). Hence, they might be false-positives of the cobas assay: The cobas assay showed that two cases (case number CT-120 and CT-135) were positive for L858R, and another two cases (CT-003 and CT-156) positive for Ex20Ins, and additional three cases (CT-040, CT-063, and CT-089) positive for Ex19Del. Interestingly, in CT-040 and CT-089, the cobas assay detected deletion in exon 19, whereas the CancerSCAN detected insertion (p.I740_K745dup, c.2217_2234dup) in exon 19. By virtue of these results, treatment option was changed from EGFR-TKI to none in five cases where no additional TK activating mutation (Table 3).
The ADPS assay detected ultra-low MAFs of 0.02% and 0.06% (CT-166 and CT-214, respectively) in two samples of subset II. Meanwhile, both the cobas assay and CancerSCAN had negative results. When the raw sequencing data from CancerSCAN was examined, mutation reads in the two-point mutations (S768I and L858R) in CT-166 and CT214 were observed at low frequency, thereby increasing the possibility that the MAF was extremely low to be detected using either the cobas assay or CancerSCAN. The last subset III (CT-190) had a MAF of 0.38%, which is high enough for ADPS assay and CancerSCAN, but not for the cobas assay. Therefore, the cobas assay was the only method that did not detect it (Table 3).
In eight of 10 discrepant cases, CancerSCAN and the ADPS assay generated congruent results, thereby suggesting that the cobas assay had false-positive results. In two of 10 cases, only the ADPS assay detected ultra-low MAFs (0.02% and 0.06%), which were significantly lower than the LoD of the other two methods. As no other method was qualified to detect MAFs in the ranging 0.1% to 0.01%, these two cases were classified as unvalidated. These results strongly indicate that the ADPS assay detected EGFR mutations with relatively high specificity and sensitivity.
DiscussionAs the clinical efficacy of target-directed therapies has been established [15,16,26,27], accurate detection of response biomarkers can be life-saving as it can determine whether a patient receive life-saving, or unnecessary treatment. Considering the technical difficulty of detecting rare mutations in high background noise, as in fine needle aspiration (FNA) biopsy specimens and/or FFPE sections with low tumor cell contents, barely representing the whole tumor mass, the first step of assay development should focus on enhancing specificity and sensitivity.
To increase specificity, the ADPS assay adopted two strategies; enhanced function of DNA polymerase and primerprobe design. The cobas assay adopts competitive hybridization that requires WT blocker [28]. In SuperSelective PCR, the mutant-specific primer design is a key factor suppressing non-specific amplification [10,11]. However, the ADPS assay does not need WT blocker because ADPS Smart DNA polymerase improves the specificity between the primer and template. The technological bases of ADPS Smart DNA polymerase can be summarized as the following [12]: whereas WT Taq DNA polymerase helps mismatched 3′ end of the primers to bind the template, ADPS Smart DNA polymerase neutralizes this effect by site-specific mutagenesis of Arg536 and 660. Although these mutations increase specificity between primers and templates, they weaken the enzyme binding to the primer-template complex, thereby lowering amplification efficiency. Additional E507K substitution mutation induces the enzyme binding to the central region of the primer-template complex, which restores the amplification efficiency. The enhanced specificity between the primer and template is the key element in the discriminatory power of detecting rare mutations in the presence of excess amount of WT DNA templates. There are other factors contributing to the discriminatory power. First, since the short primer length or lower Tm by 2°C-4°C, has minimal influence on ADPS Smart DNA polymerase, primers for EGFR assay were designed to increase DCt between the WT and the mutant templates. Second, the primer-probe design affects the amplification of each mutation due to sequence-specific binding to the template. Thus, LoDs are dependent on target sequences (S3 Table). Third, the amplicon size was significantly shorter in the ADPS assay, which might help amplify gDNA from FFPE specimens which are often partially degraded.
In this clinical validation study, we could obtain evidence of the improved accuracy of ADPS-based assay over the assay, most probably due to increased specificity (Table 3). That is, in eight of 10 cases, both the ADPS assay and CancerSCAN did not detect any mutation, but cobas assay detected various EGFR mutations, which strongly suggests falsepositive results. Conversely, the same data indicate that the specificity of the ADPS assay could be as high as that of the NGS method. In the management of cancer patients, false-positives might lead to unnecessary application of wrong treatment that can cause serious complications and delay the proper treatment. Indeed, the ADPS assay and NGS-based CancerSCAN results congruently recommend against EGFR-TKI treatments in five patients (Table 3). These results suggest that the higher specificity of ADPS assay increased DCt value, thereby resulting in enhanced sensitivity. Indeed, in two discrepant cases, ADPS detected ultra-rare EGFR mutations with MAFs of 0.02% and 0.06%. However, these data could not be validated by NGS-based CancerSCAN whose LoD is too high to detect such low MAFs [8,9]. Although these results suggested ADPS assay have relatively higher accuracy, they do not provide definite proof, or warrant similar accuracy in studies using aspiration cytology samples where the accuracy is further confounded by intratumoral heterogeneity. In other words, unresolved questions are why EGFR MAFs are frequently lower than 0.1% in samples with over 80% cancer cellularity, and what role do the rare clones play during disease progression, and most importantly, what clinical merit ADPS assay has, if the targeted therapies against the rare clones have minimal benefits to the patients.
Our study focused on detecting EGFR mutations in patients with NSCLC using FFPET samples. However, it would be valuable to investigate the feasibility of detecting EGFR mutations in other specimen types—such as FNA or bronchoalveolar lavage fluid—that are less invasive and better reflect real-time tumor status. Previous studies have shown that EGFR mutations can be detected in these types of specimens [29,30]; however, further research is needed to estimate the clinical value of detecting EGFR mutations in these specimen types among patients with NSCLC.
The ADPS assay utilizes an ADPS Smart DNA polymerase that enhances both specificity and sensitivity. The high accuracy and technical ease with no need for a competitive blocker distinguish it from other, existing assays. With these benefits, the ADPS assay may facilitate the clinical decision-making process for appropriate treatment.
Electronic Supplementary MaterialSupplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
NotesEthical Statement This study was approved by Institutional Review Board of Samsung Medical Center with a waiver of informed consent (IRB no. 2021-04-150). Author Contributions Conceived and designed the study: Park IH, Son DS, Lee BC, Kim J. Contributed data or analysis tools: Park IH, Son DS, Choi YL, Cho M. Performed the analysis: Park IH, Choi JH, Park JE. Wrote the paper: Park IH, Son DS, Kim SH. In charge of NGS data analysis and statistical comparison: Son DS, Cho M. Provided clinical specimen and intellectual input. Jeon YJ, Kim HK, Choi YS, Shim YM, Lee J. Contributed to the collection and processing of the specimen: Kang JH, Park S. Fig. 1.Multiplex strategy to detect multiple epidermal growth factor receptor (EGFR) mutations. (A) To detect Ex19Del, Ex20Ins, G719X and L858R that have multiple variant alleles, as many allele-specific primers were designed, but with a common florescent probe. (B) EGFR mutations are grouped into seven mutation groups and the four reaction master mixes with internal controls. CFO 560, CAL Flour Orange 560 (compatible dye of HEX); F, fluorophore (red: CFO 560; green: FAM); IC, internal control; MMX, master mix; Q, quencher; QS 670, Quasar 670 (Compatible dye of CY5). 
Fig. 2.Sensitivity using genomic DNA from epidermal growth factor receptor (EGFR)–mutant cell lines. (A) Seven standard amplification curves of EGFR mutation groups. As templates, 99 ng of gDNA blend with mutant allele fraction (MAF) ranging from 25.6% to 0.1% were used. △Rn is the difference of Rn (fluorescence signal of reporter probe normalized to that of reference dye) between the experimental versus baseline signal. (B) Linearity shown in the ranges from MAF 0.1%, using the Ct value as y axis, and log MAF as x axis. 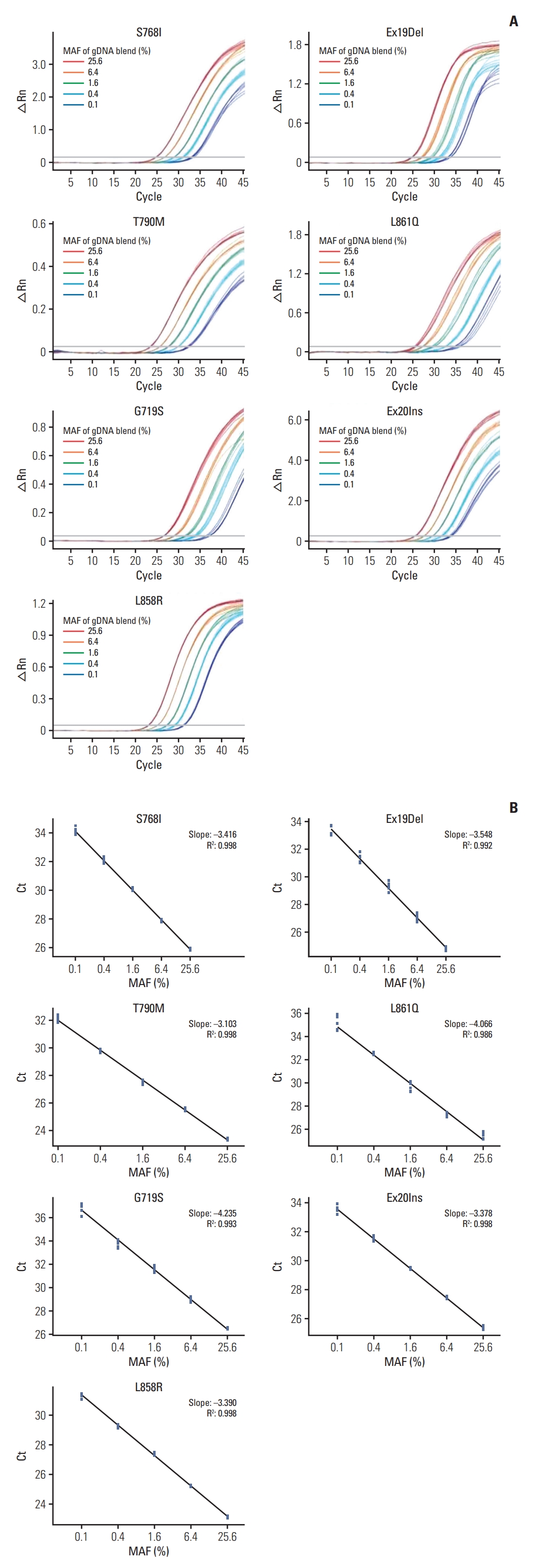
Table 1.Characteristics of specimen Table 2.Correlation between ADPS and cobas assays Table 3.Discrepancies resolved by NGS-based CancerSCAN
ADPS, allele-discriminating priming system; EGFR, epidermal growth factor receptor; NGS, next generation sequencing; TKI, tyrosine kinase inhibitor. b) Ex20Ins was negative in the ADPS assay, but Ex19Del showed a positive result in both ADPS and cobas assays, References1. Mok TS. Personalized medicine in lung cancer: what we need to know. Nat Rev Clin Oncol. 2011;8:661–8.
2. Hensing T, Chawla A, Batra R, Salgia R. A personalized treatment for lung cancer: molecular pathways, targeted therapies, and genomic characterization. Adv Exp Med Biol. 2014;799:85–117.
3. Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018;19:269–85.
4. Li X, Zhou C. Comparison of cross-platform technologies for EGFR T790M testing in patients with non-small cell lung cancer. Oncotarget. 2017;8:100801–18.
5. Nakamura H, Koizumi H, Sakai H, Kimura H, Miyazawa T, Marushima H, et al. Accuracy of the cobas EGFR mutation assay in non-small-cell lung cancer compared with three laboratory-developed tests. Clin Lung Cancer. 2018;19:170–4.
6. Syed YY. therascreen(R) EGFR RGQ PCR Kit: A companion diagnostic for afatinib and gefitinib in non-small cell lung cancer. Mol Diagn Ther. 2016;20:191–8.
7. Kim SS, Choi HJ, Kim JJ, Kim MS, Lee IS, Byun B, et al. Droplet digital PCR-based EGFR mutation detection with an internal quality control index to determine the quality of DNA. Sci Rep. 2018;8:543.
8. Shin HT, Choi YL, Yun JW, Kim NK, Kim SY, Jeon HJ, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun. 2017;8:1377.
9. Lee C, Bae JS, Ryu GH, Kim NKD, Park D, Chung J, et al. A method to evaluate the quality of clinical gene-panel sequencing data for single-nucleotide variant detection. J Mol Diagn. 2017;19:651–8.
10. Huang JK, Fan L, Wang TY, Wu PS. A new primer construction technique that effectively increases amplification of rare mutant templates in samples. BMC Biotechnol. 2019;19:62.
11. Kramer FR, Vargas DY. SuperSelective primer pairs for sensitive detection of rare somatic mutations. Sci Rep. 2021;11:22384.
12. Lim Y, Park IH, Lee HH, Baek K, Lee BC, Cho G. Modified Taq DNA polymerase for allele-specific ultra-sensitive detection of genetic variants. J Mol Diagn. 2022;24:1128–42.
13. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–46.
14. Sequist LV, Yang JC, Yamamoto N, O’Byrne K, Hirsh V, Mok T, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327–34.
15. Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113–25.
16. Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N Engl J Med. 2020;382:41–50.
17. Kancha RK, von Bubnoff N, Peschel C, Duyster J. Functional analysis of epidermal growth factor receptor (EGFR) mutations and potential implications for EGFR targeted therapy. Clin Cancer Res. 2009;15:460–7.
18. Lee CK, Wu YL, Ding PN, Lord SJ, Inoue A, Zhou C, et al. Impact of specific epidermal growth factor receptor (EGFR) mutations and clinical characteristics on outcomes after treatment with EGFR tyrosine kinase inhibitors versus che-motherapy in EGFR-mutant lung cancer: a meta-analysis. J Clin Oncol. 2015;33:1958–65.
19. Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28 Suppl 1:S24–31.
20. Hsu WH, Yang JC, Mok TS, Loong HH. Overview of current systemic management of EGFR-mutant NSCLC. Ann Oncol. 2018;29:i3–9.
21. Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376:629–40.
22. Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–42.
23. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8.
24. Huang YH, Tseng JS, Hsu KH, Chen KC, Su KY, Yu SL, et al. The impact of different first-line EGFR-TKIs on the clinical outcome of sequential osimertinib treatment in advanced NSCLC with secondary T790M. Sci Rep. 2021;11:12084.
25. Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–9.
26. Yang JC, Sequist LV, Geater SL, Tsai CM, Mok TS, Schuler M, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015;16:830–8.
27. Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–67.
28. U.S. Food and Drug Administrastion. FDA summary of safety and effectiveness data (SSED). Premarket approval (PMA)
No. P120019. Cobas EGFR Mutation Test v2. Silver Spring, MD: U.S. Food and Drug Administration; 2018.
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||