Cytoplasmic Trapping of CXCR4 in Hepatocellular Carcinoma Cell Lines
Article information
Abstract
Purpose
The chemokine receptor CXCR4 plays a role in the metastasis and progression of a broad range of malignant tumors; however, its influence on hepatocellular carcinoma (HCC) is not well defined. Thus, we analyzed the expression of CXCR4 and its functions in HCC cell lines in vitro.
Materials and Methods
Five HCC cell lines (HepG2, Hep3B, SK-HEP-1, NCI-H630 and PLC/PRF5) were investigated. The CXCR4 expression was analyzed by RT-PCR, Western blotting, flow cytometry and immunofluorescence staining. In addition, the effects of stromal cell-derived factor-1 (SDF-1) on the migration, proliferation and survival of the cells were investigated, as well as the SDF-1-induced phosphorylation of signaling molecules.
Results
All five cell lines had abundant CXCR4 in their cytoplasm, whereas a cell surface CXCR4 expression was only detected in a very small population of PLC/PRF5 cells. In contrast, SDF-1 bound to all the cells. SDF-1 induced the phosphorylation of AKT and ERK1/2 in the PLC/PRF5 cells and the phosphorylation of Stat3, AKT and ERK1/2 in the Hep3B cells. Nonetheless, SDF-1 did not induce migration or proliferation in any of the cells, nor did it rescue the cells from serum deprivation-induced apoptosis. Recruitment of CXCR4 from the cytoplasm to the cell surface was not elicited by dexamethasone, proinflammatory cytokines or VEGF. Hypoxia increased both the cytoplasmic and cell surface expressions of CXCR4 in only the PLC/PRF5 cells.
Conclusions
CXCR4 is trapped in the cytoplasm and it is not recruited to the cell surface by standard extrinsic stimuli in the majority of HCC cell lines, and the result of this is a negligible response to SDF-1.
INTRODUCTION
Chemokine stromal cell-derived factor-1 (SDF-1) is constitutively expressed in bone marrow stromal cells, and it induces the migration and homing of hematopoietic stem/progenitor cells via its G protein-coupled receptor CXCR4 (1). Disruption or blockade of the SDF-1/CXCR4 axis results in the mobilization of hematopoietic stem/progenitor cells into the peripheral blood (2).
Cancer cells also express CXCR4 and they respond to SDF-1, resulting in cellular trafficking that leads to invasion and metastasis (3-5). The serum levels of SDF-1 are elevated in patients with certain types of malignancies such as multiple myeloma (6), and this suggests that SDF-1 is involved in the development or progression of disease. SDF-1 stimulates cancer cell proliferation (4,7-9) and it protects cancer cells against apoptosis (8). In addition, SDF-1 plays an important role in tumor neoangiogenesis, and blockade of the SDF-1/CXCR4 axis attenuates tumor growth (10). These observations raise the possibility that modulation of the CXCR4 expression in cancer cells or the SDF-1 production in target organs can have an influence on cancer cell biology and the course of disease.
It was previously reported that many hepatocellular carcinoma (HCC) cell lines and the primary cancer cells in a group of patients with HCC expressed CXCR4 on their surface (11-13), that they respond to SDF-1 (12,13), and that the SDF-1/CXCR4 axis is involved in the progression of disease (13,14). However, a number of reports have contradicted this notion (15,16). As a step to clarify the role of CXCR4 in HCC, we investigated the expression status and function of CXCR4 in HCC cell lines in vitro and we showed that CXCR4 is trapped in the cytoplasm in the majority of cells, rather than being expressed on the cell surface, which results in negligible responses to SDF-1.
MATERIALS AND METHODS
1) Cells and reagents
The human hepatocellular carcinoma cell lines HepG2, Hep3B, SK-HEP-1, NCI-H630 and PLC/PRF5 were purchased from American Type Culture Collection (ATCC, Manassas, VG) and these cell lines were cultured in RPMI1640 and Dulbecco's Modified Eagle Medium (Gibco BRL Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Gibco). The human leukemia cell line MO7e was cultured in Iscove's modified Dulbecco's medium (IMDM; Gibco) supplemented with 10% FBS and 20 ng/ml granulocyte, macrophage colony-stimulating factor (Lucky Bioscience, Daejeon, Korea). The human breast cancer cell line MCF-7 was cultured in Dulbecco's Modified Eagle Medium (Gibco) supplemented with 10% FBS. Interferon-gamma (IFN-γ), tumor necrosis factor-alpha (TNF-α), transforming growth factor-beta1 (TGF-β1), vascular endothelial growth factor (VEGF) and SDF-1 were purchased from R&D Systems. Dexamethasone and pertussis toxin (PTX) were purchased from Sigma Chemical Co. (St. Louis, MO).
2) Flow cytometry
To detect CXCR4 on the cell surface, a total of 1×104 to 1×105 cells were incubated with phycoerythrin (PE)-conjugated monoclonal antibody (clone 12G5; BD Pharmingen, San Diego, CA) in phosphate-buffered saline (PBS) that contained 0.1% bovine serum albumin (BSA; Gibco) at 4℃ for 30 min, and then the cells were analyzed. Isotype-identical antibodies served as controls. To detect cytoplasmic CXCR4, the cells were permeabilized with a saponin-based reagent (BD Pharmingen) according to manufacturer's instructions and then they were labeled with PE-conjugated monoclonal antibody to CXCR4. Additional analysis of the cell surface expression of CXCR4 was performed using an Anti-human CXCR4-Biotin Sampler Pack™ (R&D Systems). Briefly, the cells to be used for staining with the antibody were first Fc-blocked with using treatment human IgG (Sigma) for 15 min at room temperature. The Fc-blocked cells were incubated with 5 µl of biotin-conjugated monoclonal anti-CXCR4s (clones 12G5, 44708, 44716, and 44717) for 30 min at 4℃. After washing twice with PBS, the cells were resuspended in PBS and then 5 µl avidin-fluorescein isothiocyanate (FITC) was added. After 30-min incubation at 4℃, the cells were washed twice with PBS and then they were analyzed. Isotype-identical antibodies served as controls (IgG2A for clones 12G5 and 44708; IgG2B for clones 44716 and 44717). To detect CXCR7 on the cell surface, the cells were incubated with allophycocyanin (APC)-conjugated monoclonal antibody to CXCR7/RDC-1 (clone 11G8; R&D Systems) at 4℃ for 30 min and then the cells were analyzed. The binding of SDF-1 to the cell surface was analyzed using human SDF-1α biotin conjugate (R&D Systems). Briefly, the cells to be stained were pretreated with purified human IgG for 15 min at room temperature for fc-blocking. Ten µl of biotynlated SDF-1α was added to 25 µl of cells suspended in PBS (1×105 Fc-blocked cells) in a 12×75 mm tube. After the incubation for 60 min at 4℃, 10 µl of avidin-FITC was added to each tube. The reaction mixture was incubated for a further 30 min at 4℃ in the dark, it was washed twice with buffered-saline protein solution (RDF1) provided by the manufacturer and then the cells were resuspended in 200 µl of RDF1 for final cytometric analysis. The specificity of the SDF-1α-biotin reaction was tested in parallel experiments. Briefly, 20 µl anti-human SDF-1α blocking antibody and 10 µl SDF-1α-biotin was mixed with the cells in a separate tube and this was incubated for 15 min at room temperature. The reaction was then allowed to proceed as described above. To detect apoptosis, the cells were stained with FITC-conjugated annexin V (BD PharMingen), according to the manufacturer's instructions, and then the cells were analyzed. Flow cytometric analysis was performed using a Coulter Elite flow cytometer (Coulter Electronics Ltd., Hialeah, FL) to detect CXCR4, SDF-1α binding and apoptosis, and a FACSCantoII flow cytometer (BD Bioscience) was used to detect CXCR7.
3) Immunofluorescence staining
Cells grown on cover slips (Paul Marienfeld Gmbh & Co KG, Lauda-Koenigshofen, Germany) were washed with cold PBS, fixed in 4% paraformaldehyde (Sigma) for 15 min at 37℃ and then they were washed three times with PBS. The cells were then incubated with a murine monoclonal anti-CXCR4 antibody (1 : 2000) (12G5; R&D Systems) for 90 min at 37℃, they were washed three times with PBS and then incubated with fluorescein-conjugated anti-mouse IgG (1 : 4000) (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) at 37℃ for 60 min. The cells were washed three times with PBS, fixed, mounted on glass slides with PBS and observed under a laser scanning confocal microscope (Olympus Corp., Lake Success, NY).
4) Cell proliferation assay
The effects of SDF-1 on cellular proliferation were measured using a colorimetric assay kit (CCK-8 assay kit; Dojindo Laboratories, Japan) according to the manufacturer's instructions. Briefly, 5×103 cells were seeded onto 96-well plates that were covered with 100 µl serum-free X-VIVO medium (BioWhittaker, Walkerville, MA) in the presence or absence of SDF-1. After 3 days incubation at 37℃ and in 5% CO2, 10 µl of a CCK-8 solution was added to each well. Two hours later, the optical density (OD) was measured using a spectrophotometer (Molecular Devices Co., Sunnyvale, CA), and the fold-increase in OD, compared with the control OD (the proliferation index), was calculated.
5) Transmigration assay
For the transmigration experiments, cells (2.0-2.5×105 cells/well) were loaded into the upper chamber of a 24-well Transwell™ plate that contained a 5-µm microporous membrane (Corning-Costar, Cambridge, MA) and they were allowed to migrate into the lower chamber over a 4-h period. The migrated cells were counted by flow cytometry, and the fold-increase in the number of migrated cells, compared with that of the control (the migration index), was calculated.
6) RT-PCR
The total RNA was prepared from cells with using Trizol reagent (Gibco) according to the manufacturer's instructions. After purification, 1 µg of the RNA was reverse-transcribed using SuperScript reverse transcriptase (Gibco) and the universal primer oligo (dT)15 (Promega, Madison, WI). In each reaction, 1 µl of cDNA was added to 24 µl of PCR buffer (Gibco) that was supplemented with 2 mM MgCl2, 0.2 µM of each primer and 1 U of Koma Taq polymerase (Koma International, Seoul, Korea). Thirty cycles of 1 min at 94℃, 45 s at 55~65℃ and 1 min at 72℃ were performed with using a GeneAmp PCR system (Perkin Elmer, Norwalk, CT). The following primers were used: human CXCR4 (sense, AAT CTT CCT GCC CAC CAT CTA CTC C; antisense, GCG GTC ACA GAT ATA TCT GTC ATC TGC C), human SDF-1 (sense, AGA ATT CAT GAA CGC CAA GG; anti-sense, AGG ATC CTC ACA TCT TGA ACC) and GAPDH (sense, CAT GTG GGC CAT GAG GTC CAC CAC; antisense, TGA AGG TCG GAG TCA ACG GAT TTG GTC).
7) Western blot analysis
Western blotting was used to detect the expressions of CXCR4 and hypoxia-inducible factor-1alpha (HIF-1α), as well as the phosphorylation of the signaling molecules. The cells were collected by centrifugation, washed in PBS and then lysed in sample buffer [62.5 mM Tris-HCl (pH 6.8), 6% (w/v) SDS, 30% glycerol, 125 mM DTT and 0.03% (w/v) bromophenol blue]. The whole-cell samples were sonicated, lysed and denatured by boiling them for 5 min. Equal amounts of protein from each sample were electrophoresed in 10~18% SDS-polyacrylamide gels and the proteins were transferred to nitrocellulose membranes (Amersham Life Science, Arlington Heights, IL). The membranes were blocked for 1 h in Tris-buffered saline (TBS) containing 5% (w/v) milk and 0.1% Tween 20 and then the membranes were incubated with mouse or rabbit monoclonal antibody overnight at 4℃. The blots were subsequently washed with TBS containing Tween 20, incubated with anti-mouse or anti-rabbit secondary antibody (Cell Signaling Technology Inc., Danvers, MA) for 2 h and then the blots were developed using West-Zol Plus (iNtRON Biotechnology, Seoul, Korea). For the phosphorylation studies, the cells were starved in serum-free medium for 16 h and then they were stimulated with cytokines. The following antibodies were used: anti-CXCR4 monoclonal antibody (Affinity Bio Reagents, Golden, CO), anti-HIF-1α polyclonal, anti-phospho-AKT polyclonal antibody (Ser473), anti-AKT polyclonal antibody, anti-phospho-ERK polyclonal antibody (Thr202 or Tyr204), anti-ERK polyclonal antibody, anti-phospho-Stat3 polyclonal antibody (Tyr705) and anti-Stat3 polyclonal antibody (all of them were purchased from Cell Signaling Technology Inc.).
8) Statistical analysis
The results are expressed as the mean±standard deviation (S.D.) of at least three experiments. The data was analyzed using Student's t-test for the paired samples. A p-value<0.05 was considered to be statistically significant.
RESULTS
1) Expression of CXCR4 in the HCC cell lines
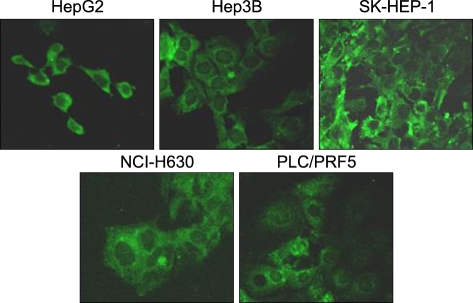
All the five HCC cell lines that were tested were positive for a CXCR4 expression, as determined by RT-PCR, whereas a SDF-1 expression was observed only in the Hep3B cells (Fig. 1A). All of the cells also produced CXCR4 protein, as was shown by Western blotting (Fig. 1B); however, flow cytometric analysis using an antibody against CXCR4 (12G5) revealed little to no cell surface expression of CXCR4 in the majority of the cells (Fig. 2A). In fact, only a small population of PLC/PRF5 cells expressed CXCR4 on their cell surface. To exclude the possibility of a low sensitivity of 12G5 antibody, the analysis was repeated using three additional monoclonal antibodies against CXCR4, and the results remained unchanged (Fig. 2B). Yet flow cytometric analysis after permeabilization of the cells revealed that all five lines contained abundant CXCR4 in the cytoplasm (Fig. 2A). This result was confirmed by immunofluorescence staining (Fig. 3).
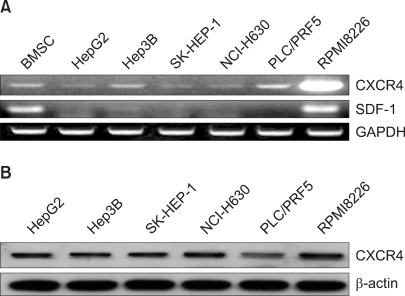
The CXCR4 expression in the liver cancer cell lines. (A) RT-PCR for CXCR4 and SDF-1 in the liver cancer cells. Bone marrow stromal cells (BMSC) and cells from the myeloma cell line RPMI8226 served as positive controls for SDF-1 and CXCR4, respectively. (B) Western blot analysis of the CXCR4 expression in the liver cancer cells.
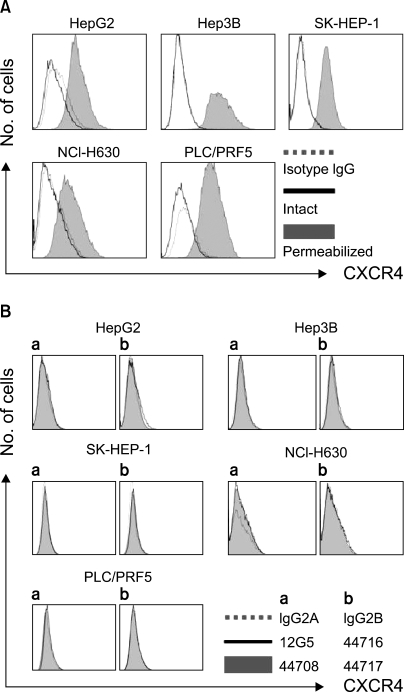
Flow cytometric analysis of the CXCR4 expression. (A) The CXCR4 expression was analyzed using antibodies against CXCR4 (12G5). The cell surface expression of CXCR4 was absent or minimal in all of the cells; however, abundant CXCR4 was detected in the cytoplasm of the cells after permeabilization. (B) The cell surface CXCR4 expression in the cells was analyzed using three additional antibodies against CXCR4 (clones 44708, 44716 and 44717). The results were the same as those produced with using 12G5.
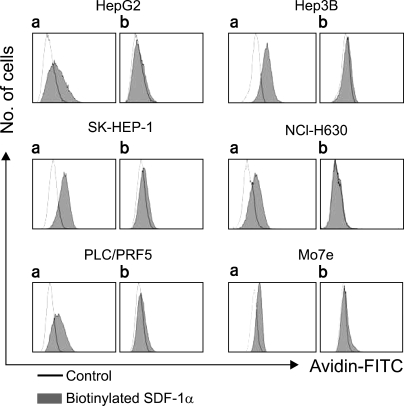
2) Binding of SDF-1 to the cell surface of the HCC cell lines
To further define the cell surface expression of CXCR4, SDF-1 binding to the cells was analyzed by performing flow cytometry with using biotinylated SDF-1α. Surprisingly, SDF-1 binding was more or less universal in all five cell lines, and this was abolished by pretreating the biotinylated SDF-1α with anti-SDF-1α blocking antibody (Fig. 4); this indicated that the binding of SDF-1 to the cells was specific.
3) SDF-1 induces the phosphorylation of signaling proteins in some HCC cell lines
Stat3, AKT and ERK1/2 were each constitutively phosphorylated to some extent in all five cell lines. SDF-1 enhanced the phosphorylation of AKT and ERK1/2 relatively late during the experimental period in the PLC/PRF5 cells (Fig. 5A), and this phosphorylation was partially inhibited by the CXCR4 antagonist AMD3100, indicating that CXCR4 responds to SDF-1 (Fig. 5C). SDF-1 also enhanced the phosphorylation of Stat3, AKT and ERK1/2 in the Hep3B cells (Fig. 5B). On comparison, the activation of AKT, MAPK and Stat3 in response to SDF-1 was not observed in the HepG2, SK-HEP-1 or NCI-H630 cells (data not shown).
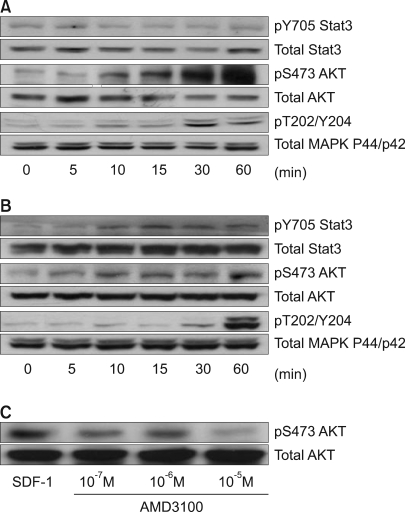
SDF-1-induced phosphorylation of signaling proteins in the liver cancer cell lines. The PLC/PRF5 cells (A) and Hep3B cells (B) were incubated in the presence of 100 ng/ml SDF-1. After the indicated time periods, the cells were subjected to Western blot analysis to evaluate the phosphorylation status of Stat3, AKT and ERK1/2. (C) AMD3100 partially inhibited the SDF-1-induced phosphorylation of AKT in the PLC/PRF5 cells. The cells were incubated with 100 ng/ml SDF-1 and AMD3100 for 10 min.
4) Expression of CXCR7/RDC-1 in the HCC cell lines
Given that the SDF-1 binds to the cells, and that SDF-1 induced the phosphorylation of signaling molecules even on the surface of the CXCR4-negative Hep3B cells, we examined whether the cells express CXCR7/RDC-1 on their surface, which has recently been reported as another receptor for SDF-1 in many tumor cell lines (17). CXCR7/RDC-1 was strongly expressed in the HepG2, NCI-H630 and PLC/PRF5 cells and to a much reduced extent in the Hep3B cells, but not in the SK-HEP-1 cells (Fig. 6).

The cell surface expression of CXCR7 in the liver cancer cells. The cells were incubated with allophycocyanin (APC)-conjugated monoclonal antibody to CXCR7/RDC-1 (clone 11G8) at 4℃ for 30 min and then the cells were analyzed by performing flow cytometry. In parallel, MCF-7 breast cancer cells were analyzed as positive controls.
5) Effects of SDF-1 on the migration, proliferation and apoptosis in the HCC cell lines
SDF-1 (at concentrations up to 200 ng/ml) did not induce chemotaxis in any of the cell lines (Fig. 7A), nor did it enhance the invasion of the cells into the matrigel (data not shown). SDF-1 also did not enhance the proliferation of the cells (Fig. 7B) or protect the cells from serum deprivation-induced apoptosis (Fig. 7C).
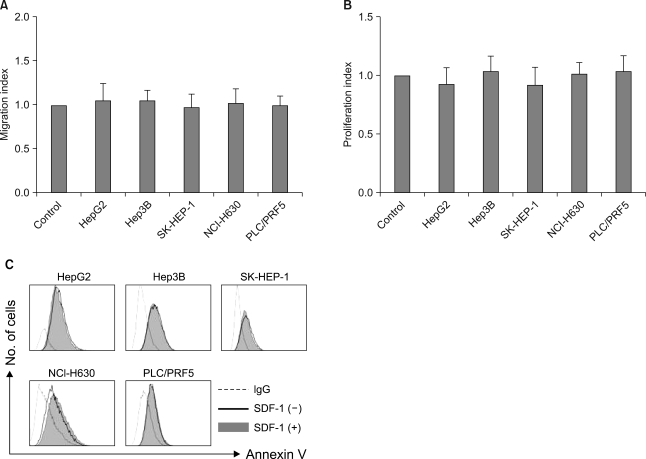
In vitro effects of SDF-1 on the liver cancer cells. (A) SDF-1 does not induce cellular transmigration. The cells were loaded into the upper chamber of a Transwell and they were allowed to migrate to the lower chamber, which contained 100 ng/ml SDF-1, over a 4-h period. The data is given as the mean±S.D. of the migration index from three independent experiments. (B) SDF-1 does not stimulate cellular proliferation. A modified MTT assay, known as a CCK-8 assay, was used. The data is given as the mean±S.D. of the proliferation index from three independent experiments. (C) SDF-1 does not rescue liver cancer cells from serum deprivation-induced apoptosis. The cells were incubated in serum-free RPMI medium in the presence or absence of 100 ng/ml SDF-1 for 24 h and then they were subjected to flow cytometric analysis.
6) Modulation of the CXCR4 expression in the HCC cell lines
We next examined the possibility that the recruitment of CXCR4 from the cytoplasm to the cell surface may be induced by extrinsic stimulation. Incubation of the cells in X-VIVO medium for up to 72 h with dexamethasone, VEGF and proinflammatory cytokines such as TNF-α, TGF-β1 and IFN-γ at various concentrations did not induce the cell surface expression of CXCR4 in any of the cells (Fig. 8). We then examined whether hypoxia up-regulates the CXCR4 expression. Incubation of the cells in 1% O2 for 16 h significantly increased the amount of CXCR4 on the cell surface and in the cytoplasm of the PLC/PRF5 cells, but this didn't occur in the other cell lines. The up-regulation of CXCR4 on the surface enhanced the transmigration of the cells in response to SDF-1 (Fig. 9).
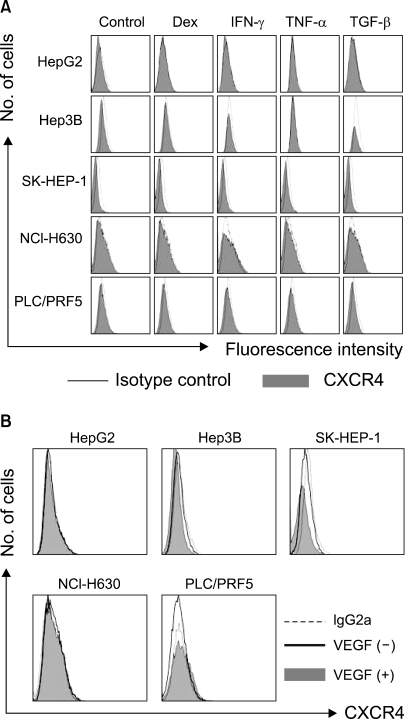
Dexamethasone and the proinflammatory cytokines (A) and VEGF (B) do not modulate the cell surface expression of CXCR4 in the liver cancer cells. The cells were incubated with 10-5 M dexamethasone, 10 ng/ml IFN-γ, 10 ng/ml TNF-α, 10 ng/ml TGF-1 or 50 ng/ml VEGF in serum-free X-VIVO medium for 24 h and then they were subjected to flow cytometric analysis for CXCR4.

Hypoxia up-regulates the CXCR4 expression in the PLC/PRF5 cells. To induce hypoxia, the cells were incubated in serum-free X-VIVO medium at 1% O2 for 16 h. (A) The cell surface expression of CXCR4 in the five liver cancer cell lines that were subjected to normoxia and hypoxia. (B) The cell surface expression of CXCR4 was enhanced in the PLC/PRF5 cells by hypoxic incubation. (a) Isotype control. (b) Normoxia. (c) Hypoxia. (C) The Western blotting showed the induction of the HIF-1α expression by hypoxia in the PLC/PRF5 cells. (D) Flow cytometric analysis after permeabilization revealed that hypoxia enhanced the cytoplasmic expression of CXCR4. (E) Hypoxia induced the transmigration of PLC/PRF5 cells in response to SDF-1. The data is given as the mean±S.D. of the migration index from three independent experiments. *p<0.05.
DISCUSSION
In this study, we showed that CXCR4 is trapped in the cytoplasm in the majority of the HCC cell lines. It has been reported that several HCC cell lines, including HepG2 and Hep3B, express CXCR4 on the cell surface (11,13), yet we were unable to detect CXCR4 on the cell surface of these cells. The expression of CXCR4 in HCC cell lines has been studied by performing a couple of experimental techniques, such as flow cytometry, immunohistochemical staining and SDF-1 binding assay. There have been significant disparities in the degree of cell surface CXCR4 expression even for the same HCC cell lines. For example, both very strong and minimal cell surface expressions in HepG2 cells were observed on the flow cytometric analysis with using the same clone of anti-CXCR4 antibody, as was reported by Mitra et al (11) and Sutton et al (13), respectively. In addition, SDF-1 binding to the cells was markedly apparent, even though the cell surface CXCR4 expression was minimal on the Hep3B and HepG2 cells (13). Membranous staining of CXCR4 was not evident in these cells in a study that used immunohistochemical staining (14). We found that CXCR4 was expressed on the cell surface in only one (PLC/PRF5) of the five cell lines, based on performing flow cytometry with using four different antibodies against CXCR4. These results indicate that CXCR4 is absent or minimally present on the surface of the HCC cells that were examined in this study. This notion is further supported by the fact that SDF-1 did not induce in vitro migration or invasion of the cancer cells, which is also in line with the results of previous studies (11,13,14). Indeed, immunohistochemical staining for CXCR4 in primary human HCC specimens indicated a predominantly cytoplasmic localization of CXCR4, and an additional weak membranous or nuclear localization in a few specimens (13,14), which is in good agreement with the results of our study with using five cell lines. It has been reported that SDF-1 binds to HepG2 and Hep3B cells (13). We also found that SDF-1 was able to bind to the cell surface in all five cell lines. Taken together, the possibility is raised that these cells have additional receptors for SDF-1.
The biological implications of the cytoplasmic trapping of CXCR4 in HCC cells are unclear. In the present study, we did not observe the cell surface expression of CXCR4 in the majority of HCC cells by performing flow cytometry; however, we cannot rule out the possibility that a very small amount of CXCR4 is present on the cell surface and that it is simply below the limit of flow cytometric detection. CXCR4 is regulated via desensitization and internalization after SDF-1 binding (18) and by transcriptional regulation (19). Constitutive and ligand-induced internalization of CXCR4 in response to autocrine SDF-1 has been observed in hematopoietic progenitor cells (12). However, the latter process does not seem to operate in liver cancer cells, as most liver cancer cells do not produce or secrete SDF-1 (15,16).
SDF-1 induced the phosphorylation of several signaling proteins in PLC/PRF5 cells via a mechanism that was partially inhibited by the CXCR4 antagonist AMD3100, indicating that the CXCR4 in the cells was functionally active. But this occurrence was not translated into migration, which may be attributed to the small portion of cells that expressed CXCR4 on their surface. It is unexpected and puzzling that SDF-1 should induce the phosphorylation of signaling molecules in Hep3B cells, which do not express CXCR4 on their surface, although they have abundant CXCR4 in the cytoplasm. SDF-1 has recently been shown to bind to CXCR7 and to transmit signals in many cell types (20). We found that many HCC cell lines express CXCR7, including the Hep3B cell line. Therefore, it is quite likely that the activation of the signaling molecules induced by SDF-1 was exerted via CXCR7. It has been shown that CXCR7 and CXCR4 play different roles in response to SDF-1 in renal progenitor cells, that is, adhesion to endothelial cells for the former and migration for the latter, respectively. (21). Based on this differential response to SDF-1, we can speculate that SDF-1 did not induce migration of Hep3B cells even though it induced the activation of signaling molecules via CXCR7. Another possible explanation is that the cells minimally express CXCR4 on their surface as mentioned above. The syndecan family may be involved in this phenomenon. Syndecan-4 has been shown to form a complex with SDF-1 and CXCR4 in lymphocytes, monocytes and HeLa cells (22). Furthermore, it has been demonstrated that syndecan-4 on its own reacts directly with SDF-1 and behaves like a receptor, at least in HeLa cells.
Steroids (23) and cytokines (24) are involved in the regulation of the CXCR4 expression in various cell types. Our study shows that steroids and cytokines, including TNF-α, TGF-β1, IFN-γ and VEGF, do not recruit CXCR4 from the cytoplasm to the cell surface, indicating that the regulation of CXCR4 is at least partially cell type-specific. Hypoxia has been shown to up-regulate not only the total CXCR4 expression, but it also up-regulates the cell surface CXCR4 expression in various cell types (25). We found that hypoxia increased the cytoplasmic and cell surface levels of CXCR4 only in the PLC/PRF5 cells, resulting in an enhanced migratory response to SDF-1. These results suggest that hypoxia is not a universal inducer of CXCR4 and that the recruitment of CXCR4 from the cytoplasm to the cell surface is not an ordinary event in the majority of HCC cells. So, it is necessary to further determine whether other extrinsic or intrinsic stimuli can induce the cell surface expression of CXCR4 in HCC cells.
CONCLUSIONS
CXCR4 is expressed on the cell surface in only a small population of hepatocellular carcinoma cells. CXCR4 is trapped in the cytoplasm and it is not recruited to the cell surface by standard extrinsic stimuli in the majority of HCC cells. Further studies on the biological and clinical implications of the cytoplasmic trapping of CXCR4 are warranted.
Notes
This work was supported by grants from the Korea Research Foundation (KRF-2005-202-E00085).