Identification of Genes Involved in Liver Cancer Cell Growth Using an Antisense Library of Phage Genomic DNA
Article information
Abstract
Purpose
Genes involved in liver cancer cell growth have been identified using an antisense library of large circular (LC-) genomic DNA of a recombinant M13 phage.
Materials and Methods
A subtracted cDNA library was constructed by combining procedures of suppression subtractive hybridization (SSH) and unidirectional cloning of the subtracted cDNA into an M13 phagemid vector. Utilizing the life cycle of M13 bacteriophages, LC-antisense molecules derived from 1,200 random cDNA clones selected by size were prepared from the culture supernatant of bacterial transformants. The antisense molecules were arrayed for transfection on 96-well plates preseeded with HepG2.
Results
When examined for growth inhibition after antisense transfection, 153 out of 1,200 LC-antisense molecules showed varying degrees of growth inhibitory effect to HepG2 cells. Sequence comparison of the 153 clones identified 58 unique genes. The observations were further extended by other cell-based assays.
Conclusion
These results suggest that the LC-antisense library offers potential for unique high-throughput screening to find genes involved in a specific biological function, and may prove to be an effective target validation system for gene-based drug discovery.
INTRODUCTION
Rapid accumulation of genomic sequence information has necessitated the need to elucidate gene functions in a high throughput mode. Various methods, including DNA or protein chips, have been devised to study the expressions of a large number of genes, which has generated a vast amount of information (1~6). However, this information is limited in that it only provides differential expression profiles for genes in different tissues or cells.
Gene expression can be specifically reduced or ablated in cells following uptake of antisense molecules that are complementary to a specific mRNA sequence. Intense efforts have been made to develop antisense anticancer agents that eliminate aberrant expression of genes involved in tumor initiation and progression (7~9). The efficacy of antisense oligonucleotides (AS-oligos) has been validated in animal models as well as in several recent clinical studies (10~12). In addition, the antisense approach has been considered ideal for functional analysis of genes and for further drug target discovery (13). AS-oligos libraries have been established and employed to obtain functional data of a large number of genes. However, constructing such a library with conventional approaches can be costly and time consuming due to the exhaustive search for effective target sites (14~17). Thus, a novel approach to facilitate easy construction of an antisense library would greatly increase the efficiency of gene functionalization and drug target discovery.
A series of distinct antisense molecules with enhanced stability, having 1) a stem-loop structure (15), 2) the CMAS (Covalently-Closed Multiple Antisense) structure (18) or 3) the RiAS (Ribbon Antisense) structure (19), have previously been described by us. Both CMAS- and RiAS-oligos, having their action point to exonucleases eliminated, exhibited greatly enhanced stability in biological fluids and were found to be effective in the specific reduction of target mRNA. The results obtained with those antisense molecules with a closed and circular structure prompted us to study the utility of genomic DNA of M13 bacteriophages as antisense molecules. The antisense sequence as a part of single-stranded large circular (LC) genomic DNA exhibited a specific antisense activity in reducing target gene expression (manuscript in preparation).
M13 bacteriophage vectors can be engineered to produce single-stranded circular genomic DNA containing an antisense sequence to a specific gene. These large circular antisense (LC-antisense) constructs may provide advantages, such as enhanced stability to exonucleases, high sequence fidelity, no target site search and easy construction of an antisense library. Thus, an antisense library encompassing a large panel of LC-antisense molecules can be readily generated and used as an effective ablator of gene expression in a high throughput mode.
Hepatoblastomas (HBs) are a subset of hepatocarcinomas and represent the most frequently occurring childhood malignant liver tumors (20). To better understand the molecular background of the disease, it would be beneficial to find genes involved in growth of HBs. In the present study, a differential antisense library was constructed using subtracted cDNA prepared from hepatoblastoma tissue, which was used to screen genes involved in the growth of a hepatoblastoma (HepG2) cell line. To do this, a subtracted cDNA library was constructed by combining procedures of suppression subtractive hybridization (SSH) and unidirectional cloning of the subtracted cDNA fragments into an M13 phagemid vector. LC-antisense molecules of 1,200 member species, selected by size, were prepared on a large scale in 96-well plates. The LC-antisense library was complexed with a cationic liposome and transfected to HepG2 cells for the massive identification of genes involved in the growth of liver cancer cells. Antisense activities were reaffirmed by other cell-based assays once the number of candidate genes had been narrowed to a manageable level.
MATERIALS AND METHODS
1) Preparation of a unidirectional subtracted liver cDNA library
To clone genes that are differentially expressed in hepatoblastomas, a cDNA library was constructed according to the manufacturer's recommendations. The SSH procedure (21) was performed with the PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, CA) using cancer and normal tissues as tester and driver, respectively. The tissue samples were washed with phosphate-buffered saline (PBS) and sliced into smaller pieces. Total RNA was extracted with Tri reagent (MRC, Cincinnati, OH). Poly(A)+ mRNA was isolated with the poly(A) Quick mRNA Isolation Kit (Stratagene, La Jolla, CA) and used as the template for cDNA synthesis. To clone the cDNA inserts unidirectionally, a cDNA synthesis primer with the HindIII recognition sequence, supplied in the kit, was replaced with a primer with a XhoI recognition sequence (5'-TTTTGTACCTCGAGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3'). First-strand cDNA was synthesized with the modified primer, followed by second-strand cDNA synthesis using T4 DNA polymerase. The tester and driver cDNAs, prepared according to the protocol, were digested with RsaI, a four-base cutting restriction enzyme that yields blunt ends. The tester cDNAs were then subdivided into two groups, and separately ligated to group-specific cDNA adaptors (Adaptor 1: 5'-ACCTGCCCGG-3', complementary to a portion of 5'-CTAATACGACTCACTATAGGGCTCGAGCGGCCGCCCGGGCAGGT-3'; Adaptor 2: 5'-ACCTCGGCCG-3', complementary to a portion of 5'-CTAATACGACTCACTATAGGGCAGCGTGGTCGCGGCCGAGGT-3'). Two hybridization reactions were performed, followed by suppression PCR to amplify the differentially expressed sequences using a pair of primers (forward 5'-TCGAGCGGCCGCCCGGGCAGGT-3' and reverse 5'-AGCGTGGTCGCGGCCG AGGT-3') and the Advantage cDNA polymerase mix (Clontech, Palo Alto, CA). The subtracted cDNA pool was cloned into the NotI and XhoI restriction sites of the multiple cloning site of the pBluescript (pBS) SK (-) vector (Stratagene, La Jolla, CA) in the same direction as the LacZ gene using the standard procedure (22). The cDNA plasmid library was transformed into Epicurian Coli XL-10 Gold Ultracompetent Cells (Stratagene, La Jolla, CA) by the calcium-chloride method (22).
To determine the total number of primary transformants, 1 or 10 µl of 1 ml pilot transformation was plated onto LB ampicillin agar plates. After overnight incubation at 37℃, the number of ampicillin-resistant colonies was counted and multiplied by a factor of 1000 or 100. To determine the percentage of vectors with inserts and the average insert size, recombinant plasmids of 40 randomly selected individual clones were purified by the alkaline lysis method (22) and digested by the same restriction enzymes used in cloning. Further, to determine if unidirectional cloning had been successfully, cDNA regions of purified plasmids derived from 100 randomly selected individual transformants were sequenced from the 5' end of the (+) strand by employing the T3 primer in an automated DNA sequencer.
2) Construction of a liver-specific random gene LC-antisense library
Bacterial competent cells containing recombinant pBS SK (-) phagemids were plated on LB agar plates containing 50 µg/ml ampicillin and 50 µg/ml tetracycline and incubated at 37℃ for 16 h. Well-isolated colonies were seeded in a well of 96 deep-well plates containing 1.4 ml of 2× YT liquid media (tryptone 16 g, yeast extract 10 g and NaCl 10 g per 1000 ml)with and additional 50 µg/ml ampicillin. Cells were cultured for 7 h at 37℃ with vigorous shaking. To produce LC-antisense molecules from each phagemid, 20 µl of the bacterial culture was transferred to each well that had been prefilled with 1.4 ml of fresh 2× YT liquid media containing 9 µl of helper bacteriophages, M13K07 (New England Biolabs, Bevelry, MA). After 1 h incubation, 4.2 µl kanamycin (70 µg/ml) was added and cultured at 37℃ for 12 h. The superinfection was carried out in triplicate for each clone to maximize the yield of antisense molecules in a single purification step. Single-stranded LC-antisense molecules were purified from the culture supernatant of bacterial competent cells using QIAprep 96 M13 Kits and QIAVAC vacuum manifolds (Qiagen, Hilden, Germany) according to the manufacturer's instructions. To test both the quantity and purity, the prepared LC-antisense molecules were run on a 1% agarose gel along with control LC-molecules derived from pBS SK (-) phagemid without a cDNA insert.
3) Transfection of the LC-antisense library into HepG2 cells
To identify genes involved in the growth of liver cancer cells, antisense molecules of the liver cancer specific LC-antisense library were mixed with a cationic liposome and transfected into the liver cancer cell line, HepG2. Briefly, the cells (7×103), washed twice with Opti-MEM (Invitrogen, Carlsbad, CA), were seeded in each well of 96-well plates in 100 µl of Opti-MEM supplemented with 10% FBS (JBI, Daegu, Korea). HepG2 cells were incubated for a further 12~18 h at 37℃ in an incubator with 5% CO2. LC-antisense molecules (0.1 µg) were complexed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in a ratio of 1:3 (w/w), and the antisense molecule+liposome complexes were added to the cultured cells according to the manufacturer's instructions. The cultures were exchanged with fresh media 24 h after transfection and incubated for a further 4 days. To compare the effects of the LC-antisense molecules on cell proliferation, identical quantities of liposome alone and control DNA+ liposome complexes were also added to the cells in a different 96-well plate and simultaneously assayed. Control DNA was single-stranded phage genomic DNA derived from pBS SK (-) phagemid without a cDNA insert.
4) Cell proliferation assay
Light microscopic observation and an MTT reduction assay were performed to study the effect of antisense molecules on the proliferation of HepG2 cells. For an MTT reduction assay, cell culture media was replaced with 50 µl fresh media and 25 µl 5 mg/ml MTT reagent (3-(4,5-dimethylthazol-2-yl)-2,5-diphenyltetrazolium bromide, in PBS) (Sigma-Aldrich, St. Louis, MO). The MTT reaction was carried out in a 37℃ incubator for 4 h followed by addition of 150 µl isopropanol, containing 0.1 N HCl, and further incubation at room temperature for 1 h. The absorbance was measured at 570 nm with a SpectraMAX 190 (Molecular Devices, Sunnyvale, CA) to quantify the surviving cells. The percentage growth inhibition of the cells in each well treated with liposome alone, antisense+liposome complexes or control DNA+liposome complexes was calculated by comparing the optical density to that of a sham treatment, using the following formula: 1-(absorbance of an experimental well/absorbance of a sham control well)×100.
5) Gene identification and sequence motif search
To identify genes complementary to the LC-antisense molecules that inhibited the proliferation of liver cancer cells, recombinant phagemids obtained by alkaline lysis were sequenced from the 5' upstream of the (+) strand of cDNA inserts using the T3 primer. The revealed cDNA sequences were compared with those of the GenBank database (http://www.ncbi.nlm.nih.gov/genebank). Polypeptides deduced from the cDNA sequences were then searched for amino acid motifs using the ProfileScan Server (http://hits.isb-sib.ch/cgi-bin/PFSCAN?).
6) Cell cycle analysis
HepG2 cells (5×104) were seeded in each well of 24-well plates. Cells were trypsinized and suspended in 250 µl PBS buffer in 48 h treatments of 0.8 µg of selected LC-antisense molecules combined with 2.4 µg of Lipofectamine 2000. As controls, sham treated cells, with liposome alone or control DNA+liposome complexes, were also prepared. Cell suspensions were fixed with 750 µl 95% ethanol by vortexing and incubating for at least 1 h on ice. Cells were centrifuged, washed twice with 1 ml PBS, and treated with 5 µl RNaseA (1 mg/ml) at room temperature for 5 min. Cells were stained with propidium iodide (PI), at a final concentration of 10 µg/ml, followed by incubation at room temperature for 30 min. The cell cycle patterns were analyzed on an equal number (104) of cells by flow cytometry using a FACSCalibur System (Becton Dickinson, Franklin Lakes, NJ).
7) Apoptotic DNA fragmentation analysis
HepG2 cells (1×106) were seeded in a 6 cm-dish one day prior to antisense transfection. Eight µg of each LC-antisense species was complexed with 24 µg of Lipofectamine 2000 and added to the cells. To compare the effects of the LC-antisense molecules on apoptosis, equal amounts of the control DNA+ liposome complexes were added to the cells for control analysis. In addition, cisplatin (Sigma-Aldrich, St. Louis, MO), an apoptosis-inducing positive-control drug, was also added, to a final concentration of 16 µg/ml. Cells were lysed with a cell lysis buffer (1% NP-40, 25 mM EDTA and 50 mM Tris-HCl, pH 6.6) 48 h post-transfection. Cell lysates were treated with RNaseA (final conc. 20 µg/ml) and incubated for 3 h at 58℃. Proteinase K was then added to a final concentration of 20 µg/ml and incubated for 3 h at 37℃. The cell lysates were mixed with 2.5 vol. of ice-cold ethanol and 0.1 vol. of 3 M ammonium acetate and stored overnight at -20℃. Precipitated DNA was resuspended in 25 µl TE buffer (pH 8.0) and run on a 1.6% agarose gel.
RESULTS
1) Construction of an LC-antisense library
A library of LC-antisense molecules was constructed with an M13 phagemid vector, utilizing the salient feature of easy cDNA library construction (Fig. 1). Messenger RNA was prepared from normal liver and hepatoblastoma tissues, and differentially amplified for liver cancer specific mRNA by a subtractive hybridization method. Amplified cDNAs were unidirectionally cloned into the pBS SK (-) vector using the NotI and XhoI restriction sites, and the cDNA library was transformed into E. coli competent cells. The primary library size was determined as 2×104 cfu/ml. By performing restriction analysis, it was determined that 97.5% of the tested transformants had cDNA inserts, and the average insert size was approximately 300 bp (data not shown). When sequenced for cloning integrity using 100 randomly selected clones, 96% of the recombinant phagemids had inserts in the intended orientation (data not shown). From the cDNA library of 9,600 transformants, 1,200 clones with cDNA inserts of more than 500 base pairs were selected with a simplified procedure for plasmid preparation (23).
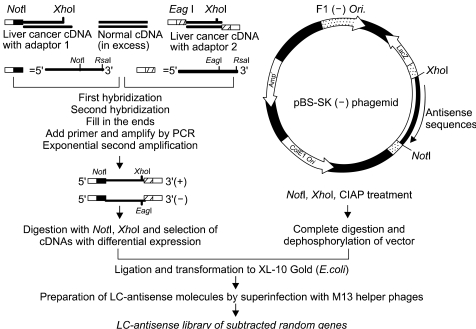
A schematic diagram for the construction of a random gene LC-antisense library. Total RNA was prepared from a pair of liver normal and cancer tissues, with both mRNA populations converted into cDNA. The tester cDNA (from cancer tissue) and driver cDNA (from normal tissue) were digested with RsaI to obtain shorter and blunt-ended fragments. Two hybridization reactions were performed, followed by suppression PCR to selectively amplify sequences overexpressed or expressed only in liver cancer cells, but not in normal liver cells. The subtracted cDNA pool was cloned into the multicloning site of the pBS SK (-) vector in the same direction as the LacZ gene. The cDNA plasmid library was transformed into E.coli competent cells. LC-antisense molecules were prepared on a large scale by superinfecting competent bacterial cells with a helper phage, and arrayed in 96-well plates.
LC-antisense molecules were purified from the culture supernatant of bacterial competent cells superinfected with helper bacteriophages. After confirming adequate purification of the antisense molecules by gel electrophoresis, the random gene LC-antisense library of 1,200 member species specific for liver cancer was arrayed in thirteen 96-well plates for further use in functional analysis.
2) Identification of genes involved in liver cancer cell growth
LC-antisense molecules from the antisense library were transfected into HepG2 cells in order to screen genes affecting the growth of the liver cancer cell line. To examine the optimal transfection conditions in HepG2 cells, various cationic liposomes were complexed with recombinant plasmid, containing the GFP or luciferase gene in various ratios, and treated to the cells (data not shown). Based on these preliminary results, each LC-antisense molecule (0.1 µg) was complexed with a cationic liposome at a ratio of 1:3 (w/w) and transfected into 7×103 HepG2 cells in each well of 96-well plates. Fresh media was added 24 h post-transfection, and the cells incubated for a further 4 days prior to the functional analysis. Cells were inspected for morphological changes under light microscopy (Fig. 2) and the growth inhibition measured quantitatively with MTT reduction assays. Of the 1,200 antisense species, 153 (~13%) were found to have varying inhibitory degrees on the growth of HepG2 cells. In contrast, cells treated with single-stranded control DNA (without antisense insert sequences) exhibited a mild level of growth inhibition, which could also be seen in cells treated with the double- stranded DNA-liposome complex. To eliminate redundancy, cDNA clones reversely complementary to the 153 growth- inhibiting LC-antisense molecules were sequenced and then searched for in the GenBank database.
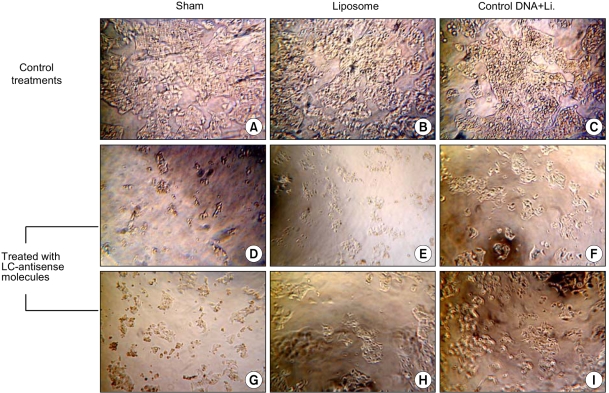
Microscopic observations after LC-antisense transfection to HepG2 cells. Growth Inhibitions of HepG2 cells were examined by light microscopy 4 days post transfection of the LC-antisense library (×200). (A)-(C); control treatments as indicated, (D)-(I); HepG2 cells treated with six different LC-antisense molecules. In this figure, the data acquired from the treatments with 6 out of 1,200 LC-antisense types are representatively shown as an example. *Li., liposome
There were 58 unique sequences out of the 153 cDNAs, which were designated as clones WGSL 1 to 58 (Table 1). From the cDNA sequence analysis, the antisense region of each clone was revealed. For instance, LCAS 3, 5, 8, 9, 10 and 23 contained the antisense sequences reversely complementary to the cDNA site 1367-1885 of WGSL 3, 21-489 of WGSL 5, 1448-2017 of WGSL 8, 421-863 of WGSL 9, 2119-2446 of WGSL 10 and 321-512 of WGSL 23, respectively. The LC-antisense molecules of the 58 clones were designated as LCAS 1 to 58. The growth inhibiting activities of the LC- antisense molecules were further confirmed by repetitive MTT assays (Fig. 3). As shown in Table 1, the GenBank annotation and motif-based searches suggested that 18 out of the 58 genes encode proteins of novel functions. The remaining 40 have an additional function to those previously characterized (e.g. protein synthesis, translation factors, metabolism and so on). These 58 genes may have functions directly or indirectly related to liver cancer cell growth.
3) Effects of LC-antisense on cell cycle progression and apoptotic induction
The 58 LC-antisense species inhibitory to cell growth were studied further to reconfirm their roles in cell growth inhibition and to understand the underlying molecular mechanisms of their inhibitory effects. To accomplish this, flow cytometric analysis was used to detect changes of the cell cycle patterns in HepG2 cells treated with the selected LC-antisense species for 48 h. When compared to the sham or control treatments employing liposome alone or single-stranded control DNA+liposome complexes, 53 (~91%) out of the 58 LC-antisense molecules exhibited varying degrees of an increasing percentage of cells with a sub-G0-G1 DNA content (Fig. 4).

Effects of LC-antisense molecules on the cell cycle progression. The LC-antisense molecules of 58 genes were transfected into HepG2 cells along with the controls. Cells were harvested 48 h post transfection. Functional analysis was perfomed on an equal number of cells (104 events) by flow cytometry after DNA staining with propidium iodide. In this figure, the data acquired from the treatments with 6 out of 58 LC-antisense types are representatively shown as an example. (A)-(C) control treatments as indicated; (D) LCAS 11; (E) LCAS 14; (F)LCAS 15; (G) LCAS 16; (H) LCAS 29; (I) LCAS 31. *Li., liposome
Whether these cell deaths, caused by the antisense treatments, reflected the induction of apoptosis were then analyzed. To do this, HepG2 cells treated with LC-antisense molecules were subjected to a DNA fragmentation assay. Twenty seven (LCAS 1, 3, 5, 8, 11, 12, 14, 15, 16, 20, 21, 22, 24, 29, 31, 33, 34, 35, 40, 41, 42, 43, 47, 49, 50, 52 and 54) out of the 58 LC-antisense species were found to cause characteristic DNA ladder formation 48 h after the transfection, indicating apoptotic progression due to the antisense treatment (Fig. 5). These results suggest that the LC-antisense library system is an effective means for en mass identification and validation of genes involved in cancer cell growth.
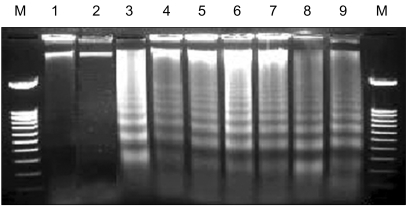
Induction of apoptotic DNA ladder formation by LC-antisense molecules. The LC-antisense molecules of 58 genes were transfected into HepG2 cells along with the controls. Genomic DNA was extracted 48 h post transfection and run on a 1.6% agarose gel. In this figure, the data acquired from treatments with 6 out of 58 LC-antisense types are representatively shown as an example. Lane M, 100 bp DNA ladder size marker; lane 1, sham treatment; lane 2, control DNA+liposome complexes; lane 3, LCAS 11; lane 4, LCAS 14; lane 5, LCAS 15; lane 6, LCAS 16; lane 7, LCAS 21; lane 8, LCAS 29; lane 9, cisplatin (positive control).
DISCUSSION
In the present study, a high throughput functional assay was developed by employing a novel large circular (LC)-antisense library. The utility of the functional genomics system was shown for en masse identification of genes involved in the growth of HepG2 liver cancer cells. Among the methods currently utilized for gene functional analysis, antisense and its related technologies are believed to be the most straightforward and efficient. To take advantage of antisense technologies, antisense libraries have been constructed using conventional AS-oligos (24). However, selection of an effective target site for each gene is time-consuming and often inconclusive (16,17). It is generally accepted that construction of an extensive AS-oligo library able to target most human genes would take years to finish, necessitating a way to overcome these problems.
It has previously been reported that ribbon antisense with a covalently closed structure is exceedingly stable and effective in eliminating target mRNA (19). Encouraged by the results obtained with the ribbon type antisense, the utility of the circular single-stranded genomic DNA of M13 bacteriophages was tested as an antisense material. It was demonstrated that large circular (LC) antisense molecules are effective in sequence-specific elimination of target mRNA (manuscript in preparation). Salient advantages of LC-antisense are the easy construction of an antisense library, the lack of need for a target site search and better antisense activity. LC-antisense is derived from cloned cDNA in a phagemid vector; therefore, a target-specific antisense region is generated as part of the single-stranded genomic DNA. This eliminates the process of the target site search that is usually required to find an effective antisense molecules. LC-antisense molecules are relatively stable to exonucleases because they have a covalently closed structure. In addition, an antisense library can be easily constructed by the introduction of a large number of cDNA fragments into a selected phagemid vector.
An antisense library has been established using 1,200 cDNA size selected clones from a subtracted cDNA library of liver cancer tissue. Cancer tissue-specific mRNA was enriched by PCR amplified subtraction, where mRNA overexpressed or expressed only in the cancer tissue was selectively amplified. Amplified cDNA inserts were unidirectionally cloned into a phagemid vector such that single-stranded circular DNA could be rescued as genomic DNA containing an antisense sequence as part of the recombinant phage genome. A large number of LC-antisense molecules were produced from the culture supernatant of bacterial cells containing recombinant M13 bacteriophages in a parallel mode. LC-antisense molecules were complexed with cationic liposomes, and then applied to target cancer cells. When using a random gene antisense library, functional assays with the antisense library can be performed without knowing the sequence of each library member. Nucleotide sequences of the antisense molecules can be analyzed after detecting the antisense activity. In conventional functional genomics, the identities of cDNA clones must be known before performing a functional assay because it is unrealistic to functionalize a large number of genes without knowing their identities and anticipated functions.
In the present study, HepG2 antisense transfectants were examined for growth inhibition, which can be studied easily with various methods, such as light microscopy, MTT reduction assay, flow cytometry and apoptotic DNA fragmentation assay. The functional analyses of genes can measure various phenotypic changes, such as cell morphology, proliferation, apoptosis, secretion and cell surface expression of polypeptides and responses to extracellular stimuli. Cell-based assays after antisense treatment may be carried out in a high throughput mode with various molecular biological, biochemical, immunological and cell biological techniques.
By employing the random gene LC-antisense library, genes involved in the growth of liver cancer cells were identified. These genes include those with an additional function to previously known ones and also those with novel functions. Further studies are clearly warranted to investigate the biochemical processes of the protein products of these genes. DNA and protein chip, or other technologies for functional genomics, can also be combined with the LC-antisense system for target validation of genes for eventual drug development. It should also be noted that the results obtained using an antisense library could be directly applied to the developments of therapeutic agents against the disease of concern. Therefore, the LC-antisense library system may provide unparalleled speed, cost effectiveness and analytical accuracy for the study of functional genomics and target validation.
CONCLUSIONS
An LC-antisense library was constructed and used to identify genes involved in liver cancer cell growth. When examined for growth inhibition with cell-based assays, 153 out of 1,200 LC-antisense molecules showed a growth inhibitory effect to HepG2 cells. Subsequent sequence comparison of the 153 clones identified 58 unique genes. The genes proved here could be promising candidates for therapeutic targets in the treatment of liver cancer. The LC-antisense library would provide potential for unique high-throughput screening to find genes involved in specific biological functions, and may prove to be an effective target validation system for gene-based drug discovery.
Notes
This study was supported by generous grants of the CDRC of Korean Science & Engineering Foundation (R13-2002-028-01004-0), 2002 Technology Innovation & Development Funds from the Small & Medium Business Administration, Korea, and WelGene Inc., a biotechnology company founded by Dr. Jong-Gu Park.

