Cancer-Specific Sequences in the Diagnosis and Treatment of NUT Carcinoma
Article information
Abstract
Purpose
NUT carcinoma (NC) is a solid tumor caused by the rearrangement of NUTM1 that usually develops in midline structures, such as the thorax. No standard treatment has been established despite high lethality. Thus, we investigated whether targeting the junction region of NUTM1 fusion breakpoints could serve as a potential treatment option for NC.
Materials and Methods
We designed and evaluated a series of small interfering RNAs (siRNAs) targeting the junction region of BRD4-NUTM1 fusion (B4N), the most common form of NUTM1 fusion. Droplet digital polymerase chain reaction using the blood of patients was also tested to evaluate the treatment responses by the junction sequence of the B4N fusion transcripts.
Results
As expected, the majority of NC fusion types were B4N (12 of 18, 67%). B4N fusion-specific siRNA treatment on NC cells showed specific inhibitory effects on the B4N fusion transcript and fusion protein without affecting the endogenous expression of the parent genes, resulting in decreased relative cell growth and attenuation of tumor size. In addition, the fusion transcript levels in platelet-rich-plasma samples of the NC patients with systemic metastasis showed a negative correlation with therapeutic effect, suggesting its potential as a measure of treatment responsiveness.
Conclusion
This study suggests that tumor-specific sequences could be used to treat patients with fusion genes as part of precision medicine for a rare but deadly disease.
Introduction
Nuclear protein in testis (NUT) carcinoma is an aggressive cancer driven by the rearrangement of the NUTM1 (NUT midline carcinoma, family member 1, NUT) on chromosome 15q14. NUT carcinoma (NC) is a very rare malignant tumor that can grow anywhere in patients of all ages but predominantly in children and young adults without gender predilection [1,2]. NC arises preferentially in a large thoracic mass that are associated with lymph nodes, bone or pleural metastases [1,3]. A large cohort study of 141 NC patients showed that 51% had primary tumors of thoracic origin and NC that arise within thorax was found at a more advanced stage [4]. Pathogenically, NC is frequently misdiagnosed as poorly differentiated squamous cell carcinoma, and thus, its actual prevalence, due to lack of awareness and/or misdiagnosis and possibility of underdiagnosis, is unknown [5]. However, improved diagnostic techniques and increased awareness have made it possible to identify and diagnose the disease more accurately and despite its low prevalence, NC was additionally categorized in the World Health Organization lung cancer classification in 2015 [6].
NC is diagnosed on clinical grounds by using immunohistochemistry (IHC), fluorescence in situ hybridization, and reverse transcription polymerase chain reaction (RT-PCR) assays, with next-generation sequencing (NGS) being used more recently. Compared to IHC, which is specific to the NUT protein, NGS identifies the presence and subtype of the fusion oncogene partner more accurately [7]. A recent study reported a prognostic model for NC patients based on an anatomical (thoracic or nonthoracic) site and NUT fusion type [4]. Upon analysis of a large cohort of 124 NC patients, most of the NC patients presented a fusion gene of NUTM1 fused to bromodomain-containing protein 3 (BRD4; 78%), bromodomain-containing protein 3 (BRD3; 15%), or nuclear receptor binding SET domain protein 3 (NSD3; 6%) genes, and 51% of them were found to be of thoracic origin. The report also showed that patients with NC of thoracic origin had the worst survival, regardless of the NUT fusion type, and the cases with thoracic NC and BRD4-NUTM1 (B4N) fusion showed poorer overall survival, although the difference was not statistically significant [4].
BRD4 protein plays an important role in transcription activation and elongation [8] and is highly enriched in super-enhancers that drive the expression of targets critical for the pathogenesis of cancer, such as c-MYC [9,10]. NUT is a testis-specific protein that is normally expressed in post-meiotic spermatogenic cells, where it facilitates hyperacetylation of chromatin by histone acetyltransferase and p300 in spermatids [11,12]. This results in both BRD4 and NUT functioning as super-enhancers that activate transcription when the tumor is formed by NUTM1 rearrangement. B4N is present in more than 80% of NCs [1] where it forms megadomains that drive ectopic transcription of underlying genes (e.g., MYC and TP63) [13], causes epigenetic reprogramming of cellular gene regulation, and stimulates uncontrolled cell growth [14,15]. To date, there is no established standard therapy for NC, and clinical treatment has not been successful [3,16]. Although local approaches, including complete resection of the tumor and initial radiotherapy, have significantly improved the survival of patients with NC, not all patients with locally advanced or metastatic cancer can benefit from this approach. Another treatment method that is currently being used is BET bromodomain inhibitors (BETis), acetyl-histone mimetic compounds, that target the bromodomain of BRD4 by competitively inhibiting BRD3/4 binding to chromatin thereby blocking differentiation and maintaining tumor growth [17,18]. For the treatment of NC, BETis have been shown to be effective in early studies against tumor progression and have also been tested in clinical trials, such as that of Birabresib (MK-8628), CUDC-907, and GSK525762 [2,19,20]. However, not all NC patients respond to BETis and the responders develop resistance to them and eventually relapse. For this reason, we sought to introduce a small interfering RNA (siRNA) therapy targeting the B4N fusion gene. siRNA has several advantages over conventional therapeutic agents because it has ability to specifically suppress a large set of cancer-related genes, regardless of the druggability of a therapeutic protein [21,22] and thereby it allows siRNA to be a potential treatment for undruggable tumors such as fusion gene-derived cancers [23,24].
In the present study, we explored whether the siRNA targeting the junction region of the BRD4-NUTM1 fusion gene has an anti-oncogenic effect, without affecting the expression of wild-type BRD4 and NUTM1. We also demonstrated its therapeutic efficacy in a mouse xenograft model. Furthermore, we determined the junction-specific sequence that could be used to detect circulating fusion genes in the blood of patients with BRD4-NUTM1 fusion by using droplet digital polymerase chain reaction (ddPCR).
Materials and Methods
1. Cells
NC SNU-2972-1, SNU-3178S, Ty-82 and HCC2429 cell lines were kindly provided by Prof. Kim at the Seoul National University Hospital [25] and synovial sarcoma HS-SY-II cells were provided Prof. Seo from the Department of Orthopedics in Samsung Medical Center. Four NC cells were maintained in RPMI-1640 (SH30027.01, Cytiva - HyClone Laboratories Inc., Logan, UT) and HS-SY-II cells were maintained in DMEM (SH30243.01, Cytiva - HyClone Laboratories Inc.) supplemented with 10% fetal bovine serum (16000-044, Gibco, Grand Island, NY) and 1% penicillin–streptomycin–amphotericin B (15240-062, Gibco) in an incubator maintained at 37°C with 5% CO2. All cell lines were performed short tandem repeat analysis for human cell line authentication.
2. Specimens
A total of 18 cases diagnosed with NC between 2010 and 2020 were selected on the basis of electronic medical records and a previous study [26] at the Samsung Medical Center. Of these, 17 samples were available, on which quantitative reverse transcription polymerase chain reaction (qRT-PCR) and/or NGS assays were performed, to identify the partner genes of NUTM1 fusion from formalin-fixed paraffin-embedded (FFPE) tissue blocks. Hematoxylin and eosin–stained slides were prepared on 4-μm-thick whole-slide tissue sections of FFPE samples. All samples were stained with hematoxylin and eosin.
3. Identification of partner genes for the NUTM1 fusion gene
RNA was extracted using the RNeasy FFPE Kit (catalog No. 73504, Qiagen, Hilden, Germany) from the collected NC FFPE samples with sufficient tissue remaining. The concentration and purity of the extracted RNA were checked using a Nanodrop and Qubit 3.0 Fluorometer (Q33216, Thermo Fisher Scientific, Waltham, MA). Samples were primarily subjected to qRT-PCR with BRD4 exon 11 and NUTM1 exon 3 primer pairs. The samples that were not detected in the qRT-PCR were subsequently subjected to targeted NGS assay. The primers used for qRT-PCR included BRD4 F 5′-GAAAGTTGATGTGATTGCCG-3′ and NUTM1 R 5′-CTGGTGGGTCAGAAGTTGGT-3′. For targeted NGS assay, in four cases, the libraries were prepared using the Archer library kit (SK0093, ArcherDX, Boulder, CO). We proceeded with PreSeq QC using 10× VCP primer mix (SA1026, ArcherDX) after synthesizing first-strand cDNA. The second-strand cDNA was synthesized by selecting samples with VCP CT values below 31. Unidirectional gene-specific primers were used to enrich the target regions, followed by NGS using the Illumina MiSeq platform (Illumina, San Diego, CA). In three cases, libraries were prepared using the Sureselect RNA Direct Library Kit (catalog No. G7564, Agilent Technologies, Santa Clara, CA). The libraries were sequenced using CancerSCAN (Seoul, Korea), a targeted sequencing platform developed at the Samsung Genomic Institution (http://kr-geninus.com).
4. In vitro siRNAs assays targeting the junction region of the fusion gene
To assess the effect of siRNAs targeting the junction region of the B4N fusion gene, a series of 15 siRNA candidates targeting the junction region formed by the fusion of BRD4 exon 11 and NUTM1 exon 3 were designed and modified with 3′-dTdT overhangs (S1 Table). ON-TARGETplus Human NUTM1 siRNA (L-022211-02-0005, Dharmacon, Horizon Discovery, Ltd., Lafayette, CO) was used as a positive control NUTM1 siRNA. For each sample, 50 nM siRNAs were transiently transfected using the Neon Transfection System into HCC2429 cells expressing the B4N fusion gene. After 72 hours, the representative images were acquired, and the cells were then harvested for qRT-PCR assays. To determine the effect of siRNAs targeting the junction region of the SS fusion gene, a series of 16 siRNA candidates targeting the junction region formed by the fusion of SS18 exon 10 and SSX1 exon 6 were designed and modified with 3′-dTdT overhanging sequences (S2 Table). For each sample, 50 nM siRNAs were transiently transfected into HY-SY-II cells expressing the SS fusion gene. Representative images were acquired after 72 hours of transfection, and the cells were then harvested for qRT-PCR assays.
5. RNA isolation and qRT-PCR assays
Total RNA from cell pellets was extracted using the RNeasy Mini Kit (catalog No. 74004, Qiagen), and cDNA was synthesized from 1 μg total RNA by using SuperScript III Reverse Transcriptase (18080093, Invitrogen, Thermo Fisher Scientific), according to the manufacturer’s instructions. qRT-PCR was conducted using a PRISM 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). The sequences of the primers used are listed in S3 Table. The experiments were performed in triplicate for all samples and t test was performed for statistical analysis.
6. Cell viability assay
HCC2429 or HS-SY-II cells were seeded into 6-well plates, and 50 nM siRNAs were transiently transfected into these cells. Twenty-four hours later, 5,000 cells were plated into 96-well plates. After 48 hours, the medium was replaced with 10 μL of CellTiter 96 AQ One Solution reagent (G3582, Promega, Madison, WI) with 90 μL of DMEM per well. The plate was incubated at 37°C for 4 hours in a humidified atmosphere with 5% CO2, and the absorbance was recorded at 490 nm using a SpectraMax 96-well plate reader (Molecular Devices, San Jose, CA).
7. Antibodies
NUT (1:50, NUT [C52B1] rabbit mAb, catalog No. 3625, Cell Signaling, Danvers, MA) and MYC (1:200, anti-c-MYC antibody [Y69], catalog No. ab32072, Abcam, Cambridge, UK) antibodies were used for IHC analysis, using a previously described procedure [26]. Immunoblotting was performed using antibodies against NUT (1:500, NUT [C52B1] rabbit mAb, catalog No. 3625), BRD4 (1:1,000, [E2A7X] rabbit mAb, catalog No. 13440), and MYC (1:1,000, c-MYC antibody, catalog No. 9402) (all from Cell Signaling Technology); SS18 (SYT) (1:1,000, [H-80], catalog No. sc-28698), β-actin (1:1,000, [C4], catalog No. sc-47778], and GAPDH (FL-335, catalog No. sc-25778) (all from Santa Cruz Biotechnology, Inc.), were used as reference, all under standardized procedures.
8. In vivo experiments
The experimental protocols were approved by the Animal Care and Use Committee of Samsung Biomedical Research Institute (Seoul, Korea). The procedures followed the institutional and National Institutes of Health guidelines for laboratory animal care, and animals were housed in an Assessment and Accreditation of Laboratory Animal Care International (AAALAC International)-accredited facility. The study was carried out in compliance with the ARRIVE guidelines. Tumors were generated in 6-week-old BALB/c nu/nu female mice. A total of 4×106 HCC2429 cells were inoculated subcutaneously into the left flank of the mice in 50 μL of a mixture of Matrigel and RPMI media (1:1 dilution). For in vivo experiments, Accell siRNA B4N #9 was customized in Dharmacon, Accell non-targeting pool (catalog No. D-001910-10-05, Horizon Discovery, Ltd.) and Accell cyclophilin B pool (catalog No. D-001920-10-05, Horizon Discovery, Ltd.) were purchased from Dharmacon as controls. Cells were transfected with siRNAs by using the Neon Transfection System (Thermo Scientific). When tumors reached an average volume of approximately 50 mm3, the mice were randomly divided into control, B4N siRNA (2 mg/kg), and B4N siRNA (6 mg/kg) groups, with five animals in each group. Mice were intratumorally injected with the siRNA every 3 or 4 days for 20 days in 20 μL serum-free media (B-005000-500, Horizon Discovery, Ltd.) per mouse. The tumor size was measured using calipers and recorded daily. We also injected cyclophilin B siRNA into three mice. The fluorescence was measured at 24- and 48-hour post-injection. Tumor volumes were calculated by taking the length to be the longest diameter across the tumor and width to be the corresponding perpendicular diameter, and then using the following formula: (length× width2) mm3×0.5. None of the animals died during the experimental period. The animals were sacrificed by means of CO2 inhalation or cervical dislocation at 48 hours after the last dose of treatment. The tumors were resected, fixed with 4% paraformaldehyde, and paraffin-embedded for sectioning and immunohistochemical staining with NUT antibody.
9. Cell-free RNA isolation and BRD4-NUTM1 transcript quantification in the plasma
For cell-free RNA (cfRNA) extraction, blood was collected in 8 mL EDTA blood tubes and centrifuged at 120 ×g for 20 minutes at RT. Then, 4 mL of the upper phase was transferred into new tubes and centrifuged at 360 ×g for 20 minutes at RT. The supernatant was removed to collect platelet pellets with 200 μL plasma at the bottom of the tube. The platelet-rich plasma (PRP) was stored at −80°C. cfRNA was isolated from 200 μL of PRP by using the miRNeasy serum/plasma kit (cat. No. 217184, Qiagen) following the manufacturer’s instructions. cfRNA was converted to cDNA by using Superscript III (Life Technologies, Darmstadt, Germany), and 8 μL of cDNA was used for droplet generation in a QX200 ddPCR system (Bio-Rad Laboratories, Inc., Hercules, CA). PCR amplification was performed using the following primer pairs (S2 Table), with GUSB as an internal control. The concentration of transcript copies in the 20 μL ddPCR reaction was reported automatically by QuantaSoft software, which was then used to determine the transcript copies per plasma using the formula: (copies per well×RNA elution volume)/(PCR volume×plasma volume).
Results
1. Patient demographics and characterization of the partner fused with NUTM1
Between 2017 and 2020, 18 patients with NC were identified through a retrospective collection of cases. The pathological characteristics of the patients are shown in Table 1. The median age at diagnosis was 46.5 years (range, 18 to 75 years) and 65% were male. Thirteen cases (72%) arose from thoracic origin such as the lung, trachea, pleura, and bronchus, and five cases occurred in the liver, nasal cavity, glottis, and metastatic lymph node. To determine the partner gene fused with NUTM1, qRT-PCR or NGS assays were performed using FFPE tumor samples. As expected, the majority of NUTM1 fusion partners were BRD4 (12 of 18, 66.7%), of which BRD4 exon 11 was found in 10 and BRD4 exon 14 was found in two cases, with BRD4 fused to NUTM1 exon 3 in both. NSD3-NUTM1 fusion was identified in two cases (of 18, 11%) (Fig. 1B). In four cases, the fusion partner genes fused to NUTM1 were not identified because one was not available for the test and two cases were not detected in the qRT-PCR assay. Unfortunately, in case No. 2, NUTM1 fusion gene was not detected in either the NGS or qRT-PCR assays. This is probably because the adequate amounts of tumoral DNA could not be extracted from FFPE samples for evaluation of the fusion gene.

Representative paired IHC images of NUT and MYC in NC patients and prevalence of NUTM1 fusion gene. (A) Immunohistochemical images in resected tumor tissue stained using anti-NUT and anti-MYC antibodies. Upper panel shows tumor images of patient No. 7 in Table 1, with NDS3-NUTM1 fusion gene in bronchus, which harbors squamous cells with pseudoglandular pattern. Lower panel presents tumor images of patient No. 15 with BRD4-NUTM1 fusion gene in lateral basal segment of right lower lobe. NUT fusion proteins formed large foci (speckled nuclear pattern) and colocalized with MYC in the nuclear region (400× magnification; scale bar=100 μm). (B) The prevalence of NUTM1 fusion gene according to the partner gene in 18 NC patients. The patient tumor samples were primarily analyzed using qRT-PCR with BRD4 exon 11 and NUTM1 exon 3 primer pairs. The samples that were not detected in the qRT-PCR were further subjected to NGS assay. For the most frequent BRD4-NUTM1 fusion samples, the sequence of the fusion breakpoint region was confirmed using Sanger sequencing. (C) Breakpoint sequence was confirmed in NUTM1 carcinoma cell lines. To determine the junction sequence of BRD4-NUTM1, mRNA was extracted from four cell lines (HCC2429, Ty-81, SNU2792-1, and SNU3178S) expressing NUTM1 fusion gene and verified using RT-PCR. The junction sequences in these cell lines were confirmed using Sanger sequencing. IHC, immunohistochemistry; NC, NUT carcinoma; NGS, next-generation sequencing; qRT-PCR, quantitative reverse transcription polymerase chain reaction; RT-PCR, reverse transcription polymerase chain reaction.
Considering that the NUTM1 rearrangement can activate oncogenes such as MYC, SOX2, and TP63, and that MYC has been reported as a downstream target of the BRD4-NUTM1 fusion protein [10], the NUTM1 fusion-positive samples were further explored using MYC IHC assays (Fig. 1A, S4 Fig.). 10 cases were stained with anti-MYC and the representative cases with the different partners of NUTM1 fusion (case No. 7: NSD3, case No. 15: BRD4) are presented in Fig. 1A. The expressed MYC was found to be localized in the nuclear compartment and was colocalized with NUT in all cases, regardless of the partner genes (S4 Fig.). As previously reported, our results showed that the expression of NUT fusion protein is strongly correlated with MYC expression in NC.
2. Development of an siRNA targeting the junction region of the fusion gene
NUTM1 fusion in NC is a major oncogenic driver gene, and we hypothesized that if the fusion oncogene was specifically eliminated from NC, the tumor growth and metastasis could be suppressed or delayed. To regulate an oncogenic function of the fusion by targeting the fusion-specific sequence, we preferentially identified the partner genes in cells with NUTM1 fusion using Sanger sequencing (Fig. 1C). Similar results obtained from clinical samples were observed in cell lines. BRD4-NUTM1 (B4N) fusion gene has been detected in three NC cell lines (SNU-2972-1, Ty82, and HCC2429), in which BRD4 exon 11 (NM_058243.2) and NUTM1 exon 3 (NM_001284292.2) were fused, and BRD3-NUTM1 fusion has been detected in SNU-3178S cell line (Fig. 1C). To explore whether targeting the junction region of a fusion oncogene is therapeutically useful, we designed a series of 15 siRNA candidates targeting the junction region of B4N (Fig. 2A, S1 Table). The candidate siRNAs were transiently transfected into the HCC2429 cells expressing B4N and 72 hours later, the relative mRNA level of parental genes and fusion gene was calculated using qRT-PCR (S5 Fig.). Commercial NUTM1 and scrambled control siRNAs were used as positive control and negative control. Next, B4N siRNA Nos. 6, 7, and 9 were treated to 293FT cells to test the effect of the siRNAs in normal cells, and the wild-type BRD4 mRNA level was measured with qRT-PCR. As expected, the junction target siRNA (B4N siRNA) did not have an effect on the BRD4 expression level (data not shown). As shown in Fig. 2A, we found that the relative mRNA level of B4N fusion decreased upon treatment with siRNAs Nos. 7 (p=0.01), 9 (p < 0.001), and 12 (p < 0.01), similar to that observed in treatment with NUTM1 siRNA, without a significant decrease in endogenous parent genes. siRNAs Nos. 12 and 15 also effectively inhibited B4N; however, they induced a concomitant decrease in the mRNA levels of B4N fusion and endogenous parent BRD4 as well. To further investigate whether the junction-specific siRNAs could affect the oncogenic function of B4N fusion, the three siRNA (Nos. 7, 9, and 12) was treated in the cell and the relative growth rate was measured using cell viability assay, and representative images were acquired (Fig. 2B and C). siRNA No. 6 was used as the negative control. As shown in Fig. 2B and C, when NUTM1 expression was downregulated by B4N-specific siRNA Nos. 7 and 9 (Fig. 2B), the relative cell growth was also delayed compared to the negative control (p < 0.001 in PC, Nos. 7, 9, 12) (Fig. 2C). These results were further investigated in synovial sarcoma. Synovial sarcoma is an aggressive neoplasm driven by the SS18-SSX fusion [27] and to determine whether siRNA targeting the junction region of a fusion gene has an inhibitory effect on cancer proliferation, 16 candidate siRNAs targeting the junction region of SS18-SSX (SS) were designed and experimented with HS-SY-II cells expressing SS fusion gene in the same manner as described above (S6A Fig.). As expected, a similar result was observed upon SS fusion gene-specific siRNA treatment. siRNA Nos. 3, 6, 7, 11, and 12 targeting the junction region of SS fusion gene induced a decrease in the relative level of SS fusion transcript and did not affect the parent mRNA expression (p < 0.001) (S6A Fig.). We showed that when SS fusion protein expression was downregulated by siRNAs Nos. 3, 6, and 11, the relative cell growth was delayed compared to that of the negative control (p < 0.001) (S6C Fig.). Similar to B4N fusion gene-specific siRNA, SS fusion gene-specific siRNAs Nos. 3, 6, and 11 selectively downregulated the expression of SS fusion protein, without affecting parent SS18 protein expression (S6B Fig.). SS siRNA No. 12 caused a decrease in SS mRNA level, but decreased the parent SS18 protein, rather than the SS fusion protein (S5B Fig.). Taken together, we showed that siRNAs targeting the junction region of a fusion gene could reduce oncoprotein expression and negatively regulate cellular growth without affecting parental protein expression. Therefore, we suggest that targeting the junction sequence of a fusion gene using siRNA could be a useful strategy to treat incurable diseases at the genetic level.
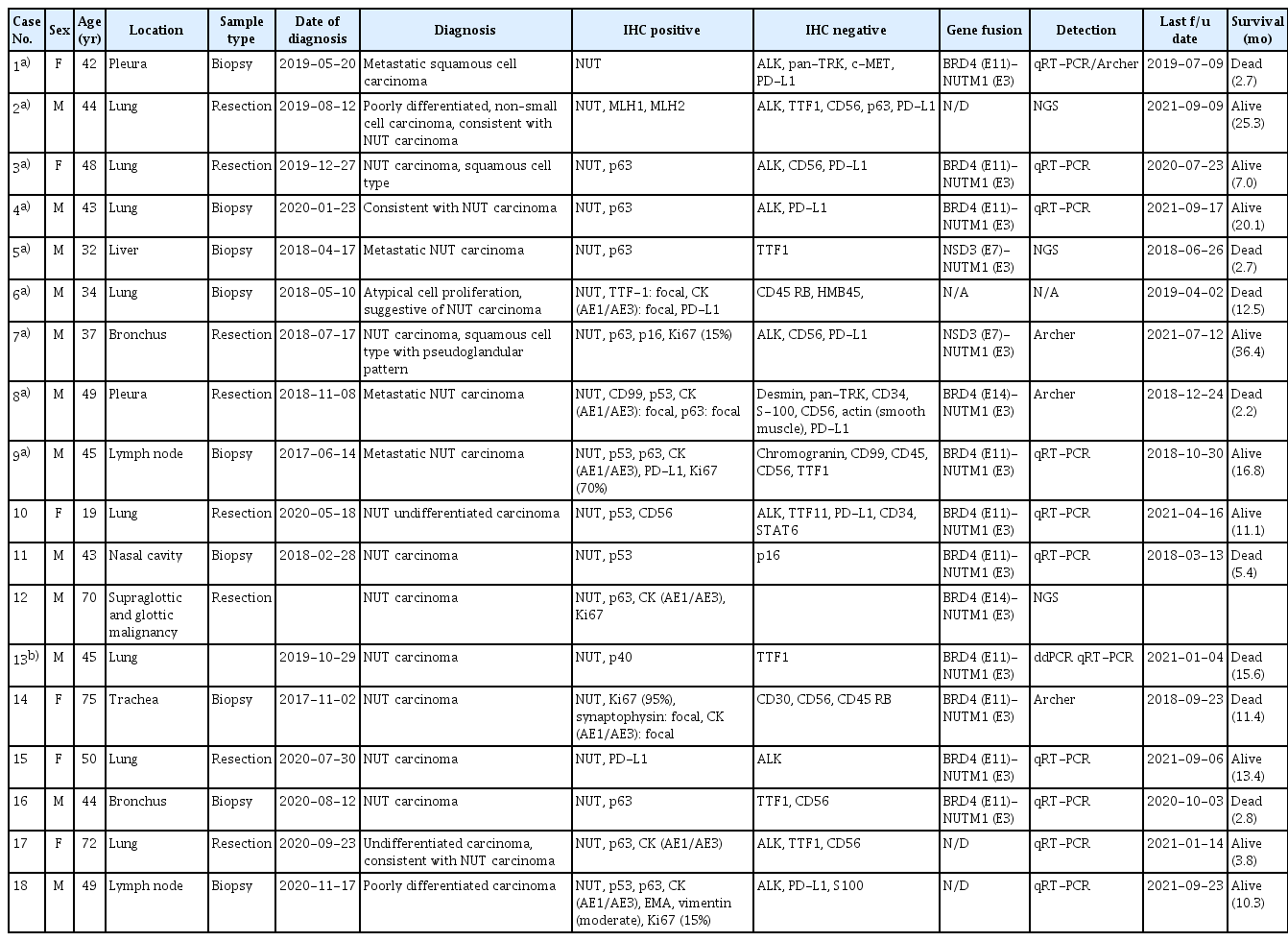
Silencing effect of siRNAs targeting the junction region of B4N fusion gene. (A) Left panel shows the scheme of junction sequence of BRD4-NUTM1 mRNA and the 15 siRNA candidates targeting the junction region, while the right panel shows the relative B4N mRNA level upon junction-specific siRNA treatment. HCC2429 cells were treated with B4N-specific siRNAs Nos. 3, 6, 7, 9, 12, and 15 at a concentration of 50 nM. After 72 hours, mRNA was extracted from the cells, and qRT-PCR was performed on the cDNA. Wild-type BRD4 level was analyzed using a primer pair for BRD4 C-terminal region, while BRD4-NUTM1 (B4N) fusion level was tested using a primer pair targeting the junction region of the fusion gene. GUSB was used as an internal control for the qRT-PCR reaction. The bar graphs represent the average values of triplicates, and error bars represent the standard deviation. (B) Immunoblots showing the expression of B4N, BRD4, and MYC in HCC2429 cells transfected with the siRNAs. The expression of B4N fusion and MYC proteins was reduced upon treatment with B4N siRNAs Nos. 7, 9, and 12 but not B4N siRNA No. 6, as compared to that in the negative control, scrambled siRNA control (indicated using a red arrowhead). (C) Cell growth assay upon B4N siRNA treatment. Cells (5×105) were seeded into 6-well plates and then treated with 50 nM siRNA. Twenty-four hours later, the cells were replated in triplicate into 96-well plates with 10,000 cells per well in 100 μL media. After 48 hours, cell growth was measured using WST assay (right panel), and representative images were acquired (left panel). p-value was determined using an unpaired Student’s t test (*p < 0.05 vs. the control group), compared with the negative control (NC) and B4N siRNA; *p < 0.05, **p < 0.01, ***p < 0.001. PC, positive control.
3. In vivo efficacy of the siRNA targeting the junction region of the fusion gene
Based on in vitro findings, we observed the effect of siRNAs targeting the junction region of the fusion gene in an in vivo experiment. For xenograft mouse experiments, B4N siRNA #9 was synthesized by Dharmacon Research with a self-delivery-modified siRNA system (Accell siRNA). HCC2429 cells were injected subcutaneously into the flanks of nude mice on day 0. When the average tumor volume reached approximately 50 mm3, the siRNAs were directly injected into the tumor areas at dosages of 2 mg/kg or 6 mg/kg, twice a week, for a total of 5 times (Fig. 3A). The localized delivery of the siRNA to a tumor site was validated using Accell cyclophilin B control siRNA, which is a highly reliable positive control for delivery and RNAi efficiency (Fig. 3B). As shown in Fig. 3C, tumor regression was observed in the B4N siRNA No. 9 treatment groups, regardless of the treatment concentration. In the 2 mg/kg treatment group, a statistically significant decrease was observed, as compared to that in the control group (p=0.024). Although not statistically significant, there also was a decrease in tumor size in the 6 mg/kg group. To determine B4N expression in xenograft tumors, the tumors were analyzed using qRT-PCR and Western blot (Fig. 3D and E). The tumors showed that B4N mRNA levels significantly decreased in the siRNA treatment groups, as compared to the control group (Fig. 3D); in addition, B4N protein expression was also reduced upon treatment of the tumors with siRNA (Fig. 3E, arrowhead).
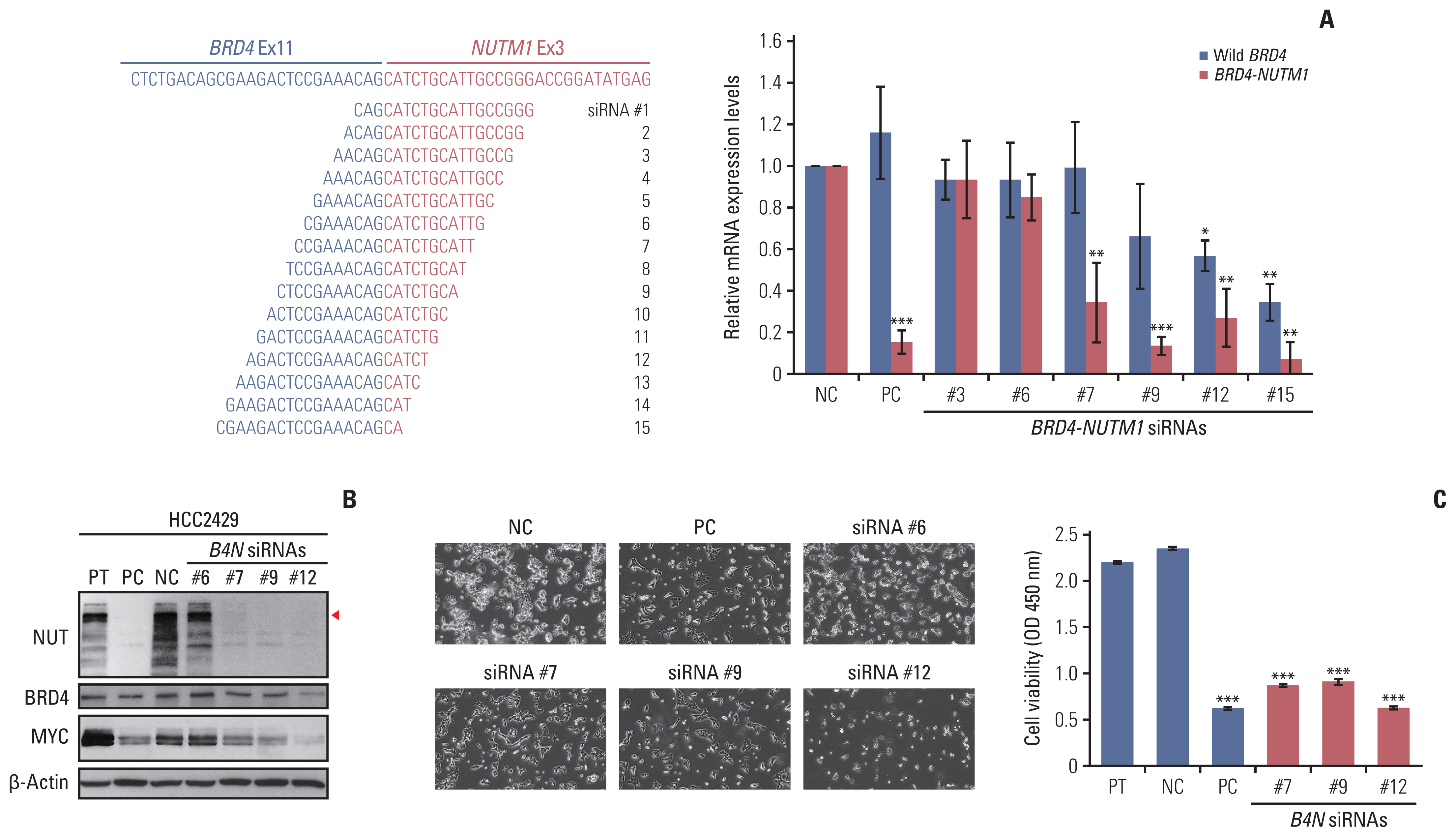
The in vivo effect of siRNAs targeting the junction region of B4N fusion gene on tumor growth. (A) A subcutaneous xenograft mouse model was generated by subcutaneously inoculating HCC2429 cells (4×106) expressing B4N fusion gene in 50 μL of Matrigel and RPMI media mixture (1:1 dilution) into the left flank of BALB/c nude female mice. When the tumors reached an average volume of approximately 50 mm3, the mice were randomly divided into three groups and either left untreated or treated with 2 mg/kg or 6 mg/kg B4N siRNA No. 9. siRNA in 20 μL serum-free media per mouse was directly injected into the intra-tumoral region every 3 or 4 days for 20 days. (B) Three mice were treated with cyclophilin B siRNA, as a positive control for siRNA injection. The fluorescence was measured (red arrow) 24 hours after injection. (C) Tumor growth curves of HCC2429 cell-derived xenograft mice. Tumor volumes were calculated by taking length to be the longest diameter across the tumor, width to be the corresponding perpendicular diameter, and then using the following formula: (length×width2) mm3×0.5. Tumor volumes were measured once every 2 days until the mice were sacrificed and presented as mean±SD (n=5 mice per group). p-value was determined using an unpaired Student’s t test (*p < 0.05 vs. the control group), compared with the control groups and B4N siRNA No. 9 (2 mg/kg)-treated group. (D) The relative B4N mRNA levels in tumor tissues of B4N siRNA-treated group. The value presents the average of each group, as determined using qRT-PCR. Statistical significance was determined using an unpaired Student’s t test: *p < 0.05 vs. the control group. (E) Protein expression of NUT and β-actin in the tumor tissues of the B4N siRNA-treated mice was evaluated using western blot (upper). Quantification of protein expression form western blot using ImageJ software (lower). *p < 0.05. qRT-PCR, quantitative reverse transcription polymerase chain reaction.
4. The potential of assaying the junction sequence for predicting and monitoring cancer treatment
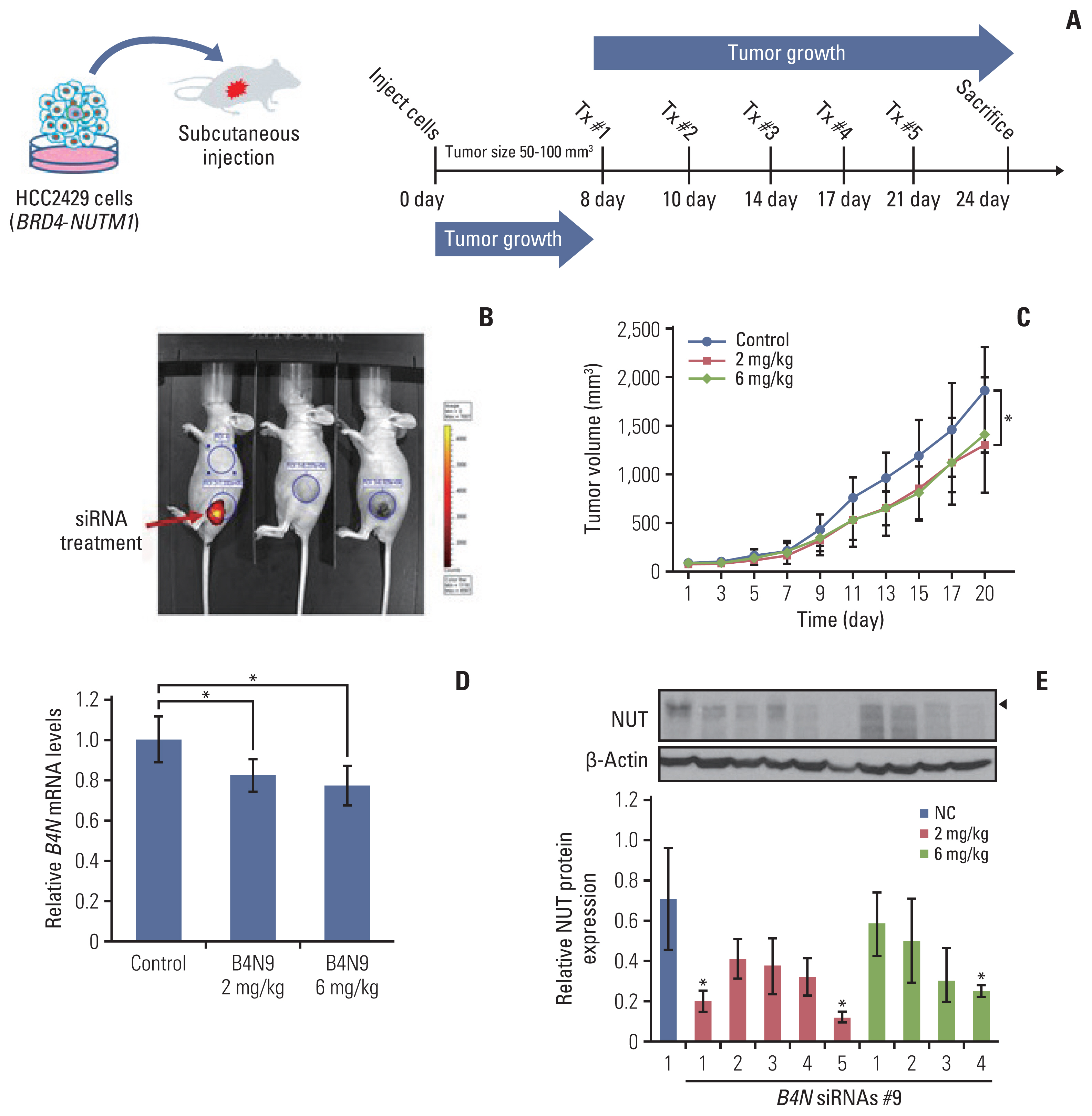
We next evaluated whether the junction sequence could be used to detect fusion oncogenes in the circulating blood of patients. This hypothesis was tested in case No. 13, a 45-year-old man, who was diagnosed with NC in the lung in November 2019 as of pT4N2M0, stage IIIB (Table 1). He underwent pneumonectomy and concurrent chemoradiation therapy following chemotherapy. Three months later, in July 2020, multiple bone metastases were detected in his case. Blood sample was the first obtained from the patient in June 2020, before radiotherapy, and subsequent blood samples were collected every 3 weeks for 28 weeks (Fig. 4B). To detect the circulating fusion oncogene in the blood, PRP was prepared from whole blood (5 mL) by means of a two-step centrifugation method (Fig. 4A). His blood was analyzed a total of nine times and 1-D fluorescence amplitude plots presented the oncogenic B4N fusion gene (S7 Fig.). To explore the association between disease progression and the number of detected droplets, the number of drops per one mL of sample was counted and plotted in Fig. 4B. The droplets in PRP were first detected when multiple tumor metastases occurred and interestingly, when the patient received radiotherapy, the plots of the B4N fusion gene immediately decreased and then increased again after a certain period of time (Fig. 4B). This showed that radiotherapy had worked for his tumor; however, unfortunately, the effect was maintained for less than 6 weeks. In this experiment, we found that the junction sequence of a gene fusion could be used to monitor the therapeutic response of patients.
Clinical usage for targeting the junction sequence of B4N fusion gene, based on the ddPCR platform. (A) ddPCR method for detecting the B4N fusion gene in PRP of an NC patient. Whole blood sample was collected and passed through a two-step centrifugation process to obtain the platelet pellet. cfRNA was extracted from the PRP sample, and cDNA was synthesized, following which ddPCR assay was performed using primer pairs against the junction sequence of B4N fusion gene. (B) The cfRNA of B4N fusion gene detected in the PRP sample. A series of blood samples were collected once a month for 8 months and analyzed with ddPCR assay, following which the number of droplets was counted and illustrated. The patient received radiotherapy and combination chemotherapy following anti-PD-1/L1 therapy. After the first sampling, the mean duration of follow-up was 21±7 days. cfRNA, cell-free RNA; ddPCR, droplet digital polymerase chain reaction; DTX, docetaxel; GEN, gemcitabine; NVB, navelbine; PD-1, programmed death-1; PD-L1, programmed death-ligand 1; Pembro, pembrolizumab; PRP, platelet-rich plasma; Ram, ramucirumab.
Discussion
NC is one of the most aggressive solid tumors, with an overall mortality exceeding 90% [11]. Over the last few years, clinical-grade small molecule BETis have been developed and administered to patients. Most inhibitors may be effective as first-line treatment; however, almost all of them tend to be short-lived and suffer from the disadvantage of responders developing resistance against them. In this study, for solid tumor patients without effective standard therapy such as those with NC, we investigated whether a tumor-specific sequence like the junction sequence of the fusion gene could be used for treatment and monitoring of the tumor, a direction toward precision and personalized medicine. For the treatment of oncogenic fusion gene-derived tumors, we applied the approach of using an siRNA against the junction sequence of B4N in NC and SS fusion genes in synovial sarcoma. Through in vitro and in vivo experiments, we showed that junction-specific siRNAs induced specific attenuation of fusion proteins, followed by cell growth arrest and decreased tumor size. In addition, by means of a ddPCR assay designed based on the junction sequence, we proved that fusion gene-specific mRNA was detected in the patient PRP, which could be used as a tool to monitor responsiveness to treatment. Our results suggested that the junction sequence of the fusion gene could be used for treatment and effective medical management in patients with fusion gene-positive cancer.
The diagnosis of NC has increased annually but given the rarity and histological similarity of NC to squamous cell carcinoma, it is still difficult. The majority (more than 70%) of NUTM1 partners in NCs belong to the BET family, including BRD4, BRD3, and NSD3, which exhibits an oncogenic function through interaction with BRD4. However, partner genes that are not related to the BET family, such as the CIC, MXD1, BCORL1, and MXI genes, have also been reported [28–30]. For this reason, we determined the partner genes of NUTM1 in our NC cases by using qRT-PCR and/or targeted NGS sequencing and showed that the majority (78%) of them, such as BRD4 (67%) and NSD3 (11%), belonged to the BRD family. Unfortunately, the fusion genes in two cases were not detected in the qRT-PCR assay, while one case was not detected in the targeted NGS assay. This may be due to an insufficient amount of tumor in the sample or a technical problem, but it does show that not all NUTM1 fusion genes are fused with the BET family. Therefore, we suggest that clinicians should plan therapeutic strategies for NC patients in cases when the NUTM1 fusion gene is detected, and the partner gene is accurately identified.
In fusion gene-derived tumor, the junction sequence is a tumor-only sequence, and it could be used as a target for treatment [23,24]. In the present study, we showed the therapeutic effects of siRNAs targeting the restricted junction region of the NUTM1 fusion gene. siRNAs were designed by spanning the junction region, following which the most effective siRNA that specifically inhibited fusion protein expression without affecting the normal protein was selected using in vitro assays. Since NUT protein is only expressed in human testis, siRNA treatment induced-toxic phenotypes are not expected in other tissues when targeting only the NUT protein. However, B4N siRNA has more precise tumor-specific targeting capabilities. Furthermore, the strategy of targeting tumor-specific sequence using siRNA is thought to be applicable to fusion-driven cancer which does not have standard treatment.
Using a mouse xenograft model, we showed that tumor growth could be moderately inhibited upon siRNA treatment. The tumor volume and NUT expression did not significantly decrease, but this may be due to the large deviation of tumor volume in different individuals because the average volume of the overall tumors has decreased as seen from the tumor growth curve in Fig. 3C. Nevertheless, this result suggests that the limited junction sequence could be a target for the treatment of patients with fusion genes that drive aberrant growth. Despite the advantages of siRNA-based technology as an effective cancer therapeutic approach, it still faces limitations in terms of efficient cellular uptake and long-term stability.
Circulating tumor DNA (ctDNA) has emerged as a blood-based biomarker for monitoring the disease status of patients. However, the usefulness of ctDNA for recurrence monitoring is limited, because tumor DNA acquires multiple genetic changes, which lead to tumor development, and thus, ctDNA is not an exact match. For this reason, the present study monitored cfRNA in blood by detecting the junction sequence of the fusion gene in the PRP sample. For all samples, cfRNA was extracted from the PRP sample and cfDNA synthesized from it by using RT-PCR was used as a template for the ddPCR assay, which is known to be more precise and sensitive, and most importantly, associated with absolute quantification. The ddPCR assay shows the amount of the oncogenic fusion gene in the patient’s blood, with detection of drop plot numbers. From this assay, we concluded that an increase or decrease in the droplet number reflects an increase or decrease of the fusion oncogene secreted from the tumor in the circulating blood. Based on our ddPCR assay, we found a decrease in fusion oncogenes in the blood post-irradiation treatment, followed by an increase in the amount of oncogene again after a certain period of time. We found that radiotherapy had a short-term effect on the patient, but the patient’s condition worsened, and he died a month after the last blood sampling. Taken together, we suggest that the junction sequences of fusion genes could be a blood-based biomarker for monitoring therapeutic responsiveness.
In this study, we demonstrate the potential of tumor-specific sequences as diagnostic tools in patients with rare cancers, which will help them to avail appropriate treatment (Fig. 5). These sequences could be used in therapeutic approaches to inhibit cancer progression and in the treatment of rare solid tumors when there is no known appropriate treatment and the tumor has oncogene-specific sequences such as fusion genes. Likewise, to increase the survival rate of fusion gene-derived cancer, despite its limited efficacy as a single agent, an siRNA targeting the junction region of the fusion gene could be used a personalized and targeted therapy, in combination with an efficient delivery platform. Although this approach may not be an ultimate solution, it will lead to better clinical outcomes in combination with the available treatment options.
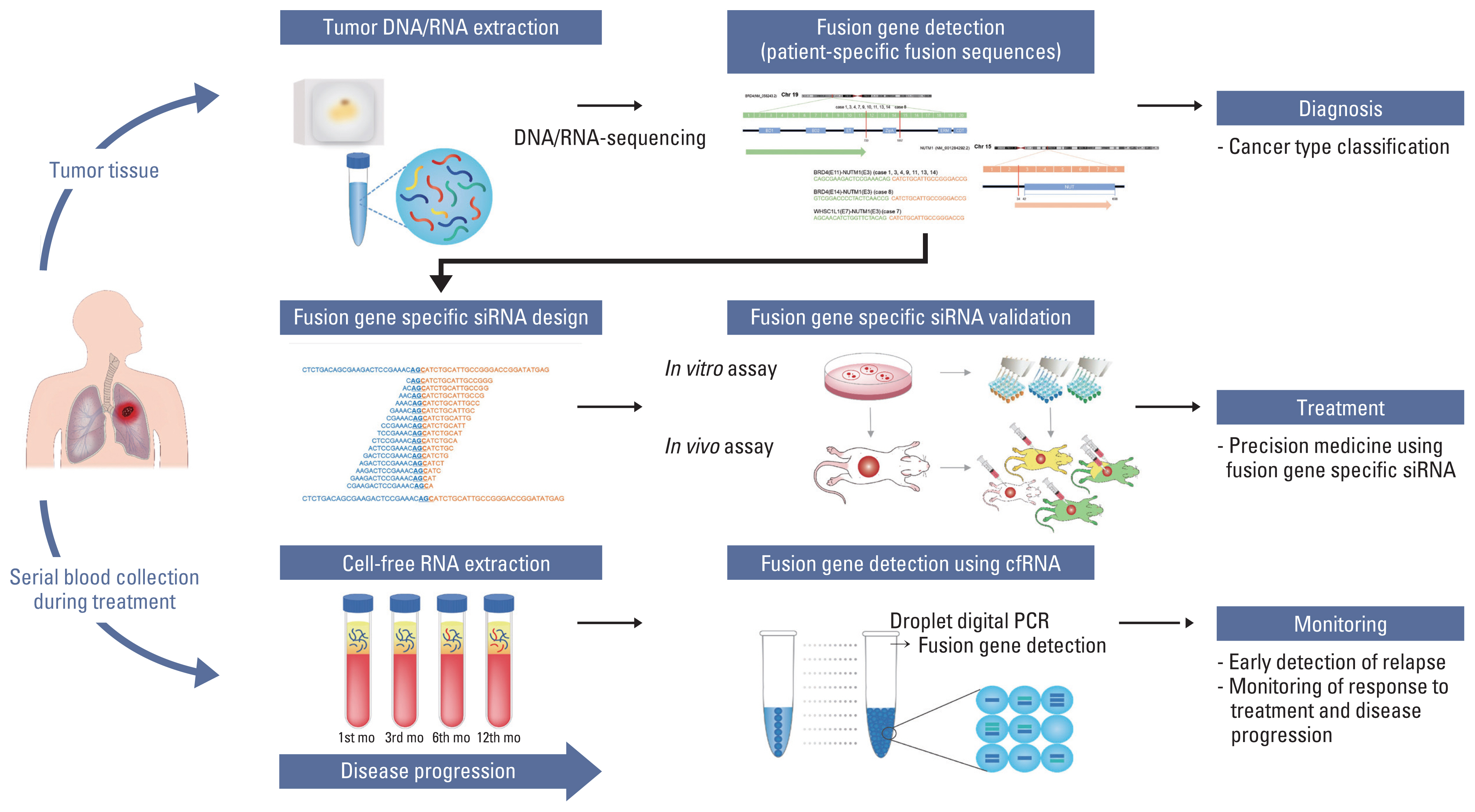
Clinical applications for fusion gene-derived cancer diagnosis and guidance of potential targeted therapies. For treatment guidance of patients with unknown solid tumors, we suggest that the tumor be sequenced and analyzed to identify the presence of tumor-specific alterations. On doing so, if a fusion gene that can be presumed to be an oncogenic driver gene is found, junction sequence of the fusion gene could be used for treatment and monitoring of therapeutic responsiveness. siRNAs targeting the junction region of the fusion gene can be developed for treatment of the patients, and cell-free RNA obtained from their blood can be used to monitor the therapeutic responsiveness. This allows for personalized treatment of individual patients, even in case of rare cancers that are ineffective with standard treatments.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
Notes
Ethical Statement
This study was reviewed and approved by the Institutional Review Board of Samsung Medical Center (approval no. 2019-05-141) and all experiments with mice conformed to the Animal Welfare guidelines and were performed in accordance with the protocols approved by the Ethics Committee.
Author Contributions
Conceived and designed the analysis: Lee MS, An S, Choi YL.
Collected the data: Sung M.
Performed the analysis: Lee MS, An S, Song JY, Jung K, Chang ES, Choi J, Oh DY.
Wrote the paper: Lee MS, An S.
Provided tumor samples: Jeon YK.
Critical comments on the manuscript: Yang H, Lakshmi C, Park S, Han J, Lee SH.
Conflicts of Interest
Conflict of interest relevant to this article was not reported.
Acknowledgments
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (Ministry of Science, ICT and Future Planning) (NRF-2021R1A2C4002158 and 2022R1A2C2006322) and SMC-SKKU Future Convergence Research Program Grant.