INTRODUCTION
More than two thirds of colorectal cancer patients undergo primary tumor resection, with one third of these going on to develop a metastatic disease that demands systemic chemotherapy (1,2). In the 40 years since the introduction of 5-FU to clinical practice, the only improvements in the modest response rate achieved with this agent have been through the modification to its administration and the use of concomitant agents that modulate 5-FU activity (3).
Considerable scope remains for improving outcomes in patients with colorectal cancer. For patients failing to respond to or having relapsed following 5-FU-based chemotherapy, some second-line chemotherapeutic agents have provided remarkable results. In the 1990s, two agents, irinotecan (CPT-11, Campto®) and oxaliplatin, were found to have activity against advanced colorectal cancer. CPT-11 has been shown to be an effective alternative to 5-FU, exhibiting a remission rate of 11~23% (4). In addition, the combination of CPT-11 and 5-FU with leucovorin has been approved as first-line chemotherapy for patients with metastatic colorectal cancer (5,6).
Evaluating the responsiveness to specific chemotherapeutic agent is considered useful, but no specific maneuver has been reported in colorectal cancer. Many reports suggest that the DNA mismatch repair (MMR) system might be involved in the tumor responsiveness to chemotherapeutic agents that target DNA (7,8). Given the incidence of MMR defects among colorectal cancers, it may be particularly relevant that MMR defects can lead to hypersensitivity to CPT-11 (8). However, the responsiveness to CPT-11 in relation to MMR status remains to be fully established.
The aim of this study was to determine the correlation between CPT-11 responsiveness and the expression of MMR protein in colorectal cancer patients.
MATERIALS AND METHODS
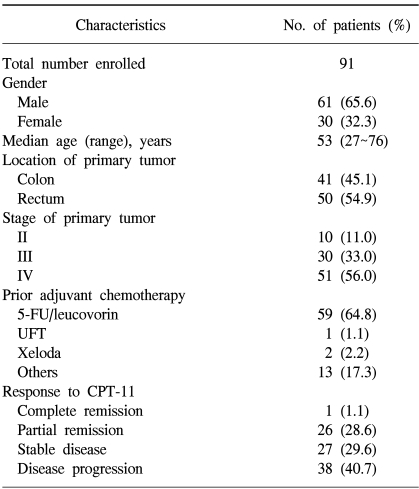
This study involved 91 patients that had received CPT-11-based chemotherapy, between 1997 and 2002, for advanced or metastatic colorectal cancer following surgery. Sixty-one patients were male and the overall mean age was 53 years (range, 27~78 years). Patients who stopped chemotherapy after the first cycle due to toxicity, economic status or incorporation were excluded. Fifty-nine patients (63.4%) had failed to respond to prior 5-FU-based chemotherapy, and 15 (16.5%) received CPT-11-based chemotherapy as an initial regimen for a metastatic disease. Of the patients with a metastatic disease, 4 underwent palliative stoma construction and the others a palliative resection of the primary tumor. Among the patients with a prior history of 5-FU based chemotherapy, 6 underwent a resection or hepatic arterial chemotherapy for a metastatic disease prior to the CPT-11-based chemotherapy. Fifty-three patients received CPT-11-based chemotherapy without exploration for a recurrence or metastasis. The median follow-up was 16 months (range, 1~32), with a mean of 5 cycles (range, 2~14) of chemotherapy administered per patient.
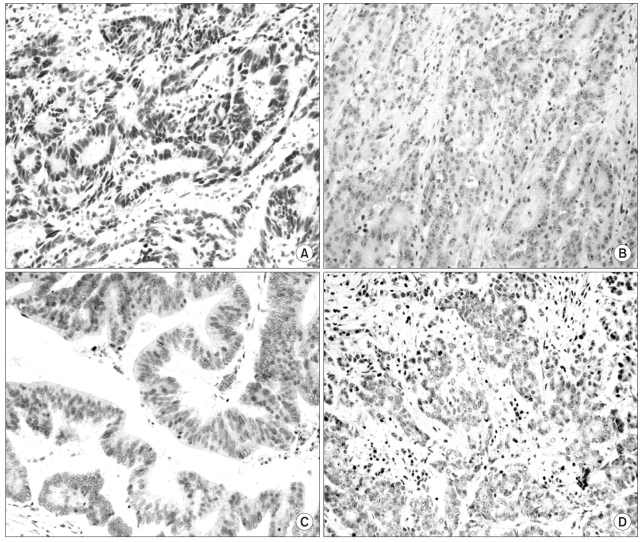
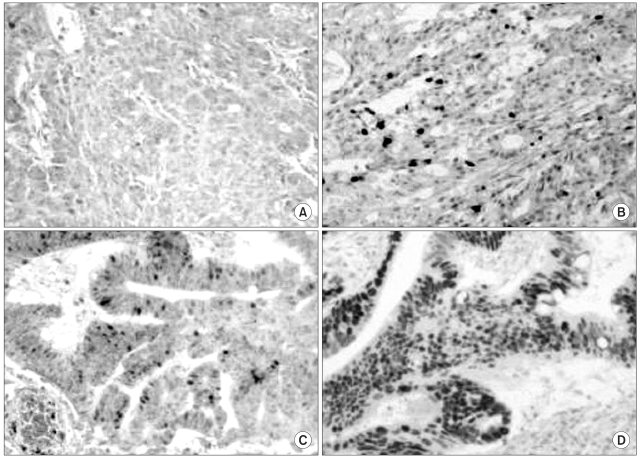
The expressions of hMLH1 and hMSH2 proteins in the tumors were examined using immunohistochemical staining. Well-preserved formalin-fixed paraffin-embedded tissue blocks from the primary cancers were selected, and blocks produced using a tissue micro-array method. Five-micrometer sections were deparaffinized and rehydrated using xylene and alcohol. Before the immunostaining, heat-induced antigen retrieval was performed by microwave oven treatment (30 min at 600 W) on the tissue sections immersed in 10 mM citrate buffer (pH 6.0). Sections were then incubated overnight at 4℃ with mouse monoclonal antibodies against hMLH1 (clone G168-728, PharMingen, San Diego, CA) and hMSH2 (clone FE11, Oncogene research products, Cambridge, MA). The p53 and CEA expressions were also measured using immunohistochemical staining. A commercially available biotin-blocking kit (Dako, Denmark) was used as the secondary detection system, and the peroxidase reaction developed using diaminobenzidine tetrahydrochloride as the chromogen. Finally, slides were lightly counterstained with Mayer Hematoxylin. Normal expressions of hMLH1 and hMSH2 were defined as the presence of nuclear staining within tumor tissues as well as within the adjacent non-neoplastic tissue, such as the mucosa. Tumors showing loss of nuclear staining were classified as having lost hMLH1 and hMSH2 (Fig. 1). For p53 expression, the nuclear staining was categorized into four grades, i.e. grade 0 for no staining or ≤10% tumor cells stained, grade 1 for >10% but ≤33.3% of tumor cells stained, grade 2 for >33.3% but ≤66.7% of tumor cells stained, and grade 3 for >66.7% of tumor cells stained (Fig. 2). The distribution patterns of CEA staining were categorized into either apicoluminal or diffuse-cytoplasmic patterns. The intensity of CEA positive staining was classified into three grades according to the standard color guide (4th ed. Dainippon Ink and Chemical Inc., Japan) (9). Two pathologists, without knowledge of the clinicopathological data, independently evaluated all the tumors.
Complete blood cell counts, blood chemistry and serum tumor marker levels were obtained prior to (baseline) and following every two cycles of chemotherapy. Tumors were evaluated by CT scans during the 2 week rest period between treatment cycles. Complete remission (CR) was defined as the complete disappearance of all clinically evident disease. Partial remission (PR) was defined as a decrease of more than 50% of the sum of the products of the largest perpendicular diameters of the measurable disease. A stable disease (SD) was defined as a 25~50% decrease in the tumor size. A progressive disease (PD) was defined as an increase in the above measurements or the appearance of new lesions (10). The response was decided on the basis of imaging studies and the opinion of an oncologist. Patients were evaluated for toxicity, which was graded using the National Cancer Institute Common Toxicity Criteria (11).
The progression-free survival was determined from the start of treatment to the time of relapse. The overall survival was calculated from the date of chemotherapy commencement to the death of the patient for any reason. Associations between clinicopathological variables and the response rate to CPT-11 were compared using t- or Fisher, s exact tests. Overall and progression-free survival curves were constructed using the Kaplan-Meier method; with the survival durations compared using the log rank test. The level of significance was set at 5% for each analysis, and all calculations were performed using the SPSS software (ver. 11, SPSS Inc., Chicago, IL).
RESULTS
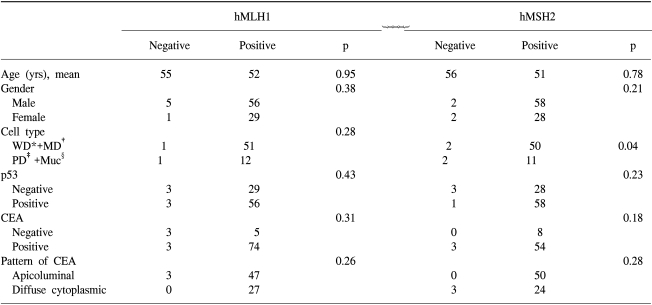
Forty-one (45.1%) of the primary tumors was located in the colon, and the remainder (50 tumors, 54.9%) in the rectum. Seventy-five patients received adjuvant chemotherapy prior to the CPT-11 therapy, and 59 of these were administered 5-FU with leucovorin-combined therapy (Table 1). There were no differences in the clinicopathological characteristics between patients with positive and negative expressions of MMR protein (Table 2).
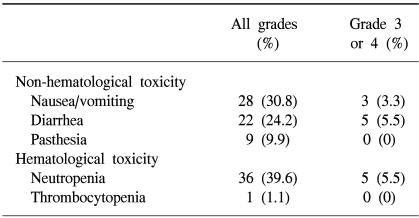
Overall, 27 patients (29.7%) showed remission, comprising one CR and 26 PR. Twenty-five patients (27.4%) were classified as SD. The response rate in patients who had previously received CPT-11-based chemotherapy as a primary treatment was 50% (8/16), while that in patients who had previously received other types of chemotherapy was 25.3% (19/75; p=0.05). The response rates according to the type of operation were no different between the R0, R1 and R2 groups. Toxicity, either hematological or non-hematological, was observed in 38 patients (41.8%), with neutropenia being the most common, encountered in 36 patients (39.6%), and grade 3 neutropenia or higher observed in 5 (5.5%). Gastrointestinal toxicities ranged from acute episodes of nausea and/or vomiting (28 patients, 30.8%) and abdominal pain, to the more troublesome toxicity of delayed diarrhea (22 patients, 24.2%), with five patients (5.5%) being hospitalized for grades 3 or 4 diarrhea (Table 3). The median survival and progression-free survival were 18 (range, 1~32) and 8 months (range, 1~16), respectively.
The expressions of hMLH1 and hMSH2 proteins were detected in 85 (93.4%) and 87 (95.6%) tumors, respectively. hMLH1 and hMSH2 were not detected in 6 (6.6%) and 4 patients (4.4%), respectively. The p53 expressions were grades 0, 1, 2 and 3 in 32 (35.2%), 8 (8.8%), 13 (14.3%) and 38 tumors (41.8%), respectively. Eight (8.8%) and 77 (84.6%) of tumors were negative and positive for CEA expression, respectively, but the expression status could not be confidently assigned in 6 samples (6.6%) due to vague staining. Of the CEA positive samples, 17 (18.7%) were weakly positive (+), 33 (36.3%) moderately positive (++) and 27 (29.7%) strongly positive (+++). Fifty CEA-positive samples (54.9%) showed apicoluminal expression, whereas 27 (29.7%) showed diffuse cytoplasmic expression.
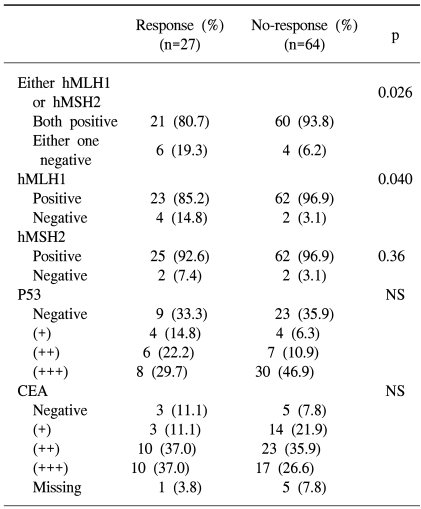
The response rates of MSI and MSS tumors were 60 and 25.9% (p=0.026), respectively. The objective rate of response was 66.7% among patients whose tumors did not express hMLH1, which was significantly higher than the 27.1% of those showing expression (p=0.04). In contrast, hMSH2, p53 or CEA expression did not correlate with the response rate (Table 4). The overall incidence of grade 3 or 4 toxicities was significantly higher in patients whose tumors showed loss of hMSH2 expression (p=0.04), while expression of hMLH1 and p53 had no correlation with toxicity.
The median overall survival in the response group was longer than that in the no-response groups (16 vs. 10 m, p=0.003) (Fig 3), as was the median progression-free survival (11 vs. 5 months, p<0.001). Survival was not affected by the expressions of hMLH1, hMSH2, p53 or CEA.
DISCUSSION
Numerous studies have reported that tumors expressing MMR proteins display different clinicopathological features, including proximal location, poor differentiation and better clinical outcome, compared to those not expressing these proteins (12,13). It is unlikely that tumors with such distinct clinicopathological features would respond similarly to the same chemotherapeutic agents. Clinical studies investigating the relationship between MMR status and chemotherapy responsiveness have stimulated some controversy (14,15). In vitro studies have shown that MMR-deficient cell lines display moderate resistance to methylating agents and low resistance to cisplatin (16,17). Using a model of colorectal cancer xenografts in nude mice, tumors not expressing MMR proteins were found to be somewhat more sensitive to CPT-11 (18). In a study analyzing 72 metastatic colorectal cancers, MSI tumors were found to have a better response rate (57.1%) to CPT-11 than MSS tumors (10.8%) (19).
In the present study, MMR gene expression was evaluated by immunohistochemical staining of tumor tissue for hMLH1 and hMSLH2 proteins. Testing tumors for the presence of underlying mismatch repair gene deficiency requires the services of a molecular genetics laboratory, and such laboratories are not readily available to most centers. Immunohistochemical analysis is a complementary method for detecting mutator phenotypes in colorectal tumors (20,21). In many studies, the sensitivity and specificity of immunohistochemical analyses of hMLH1 and hMSH2 expressions for detecting underlying mismatch repair gene deficiencies were over 90% (20,21). Therefore, immunohistochemistry appears to be a good surrogate test for the detection of MSI caused by hMLH1 and hMSH2 dysfunctions. Previously the MSI status was compared using immunohistochemical methods in 80 tumors, with two of these methods found to give identical results (22). In the present study, it was found that the response rate to CPT-11-based chemotherapy was significantly greater in patients with tumors not expressing the hMLH1 protein compared to those that did, suggesting hMLH1 protein expression may be predictive of the responsiveness to CPT-11. The molecular mechanism underlying the sensitivity of MSI tumors to CPT-11 is not completely known. CPT-11 is a derivative of camptothecin, which interferes with and stabilizes DNA topoisomerase I-cleavable complexes. This results in single-strand breaks in the DNA, which are converted to double-strand breaks and DNA damage, ultimately leading to inhibition of DNA replication and transcription (23). Stabilization of the cleavable complexes by CPT-11 treatment is accompanied by G2/M arrest and apoptosis. Along with the increase in topo-1 activity, tumor-associated deficiencies in DNA repair and cell cycle regulation, and/or inability of cancer cells to repress apoptosis, were also suggested to additionally contribute to the cancer specificity of CPT-11. The MMR system is best known for its role in post-replicative repair. Since any gene containing a microsatellite repeat is a potential target for MSI driven insertion/deletion mutations, MSI tumors accumulate widespread mutations, not only in the genes participating in tumor initiation and progression, but also in those involved in various DNA repair pathways. Recent data certainly indicate that MMR-deficient cells are able to more efficiently repair double-strand breaks by homologous recombination than MMR-proficient cells, but are also more error-prone (24). In the present study, the expressions of hMSH2, p53 and CEA did not affect the sensitivity to CPT-11. In particular, these data suggest sensitivity to CPT-11 is specific to hMLH1 rather than hMSH2 expression.
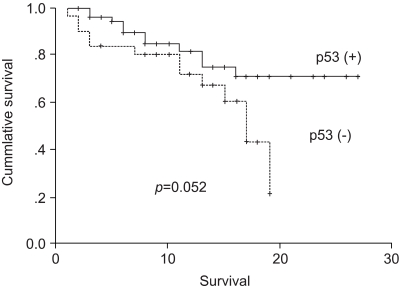
Both the median survival and progression-free survival were significantly prolonged in the responder group compared to those in the non-responder group. In addition, the survival appeared to be better in patients whose tumors did not express altered p53, although this difference did not quite achieve statistical significance (p=0.052). Most studies suggest that the presence of a p53 mutation is associated with poorer outcomes among patients with colorectal cancer (25). A meta-analysis of 28 published studies on p53 alterations reported that the presence of p53 mutations was an independent prognostic factor for overall survival. In addition, Allegra et al. recently reported that positive p53 staining was associated with a worse outcome. Out results also suggest that p53 analysis may assist in predicting survival.
It is useful to note some limitations of the present study. The enrolled patients were a heterogeneous population, making it difficult to completely understand the influence of the MMR status on the response to CPT-11. The population included patients treated with CPT-11 as a primary, and secondary regimen only. Therefore, the response rate might have been confusing. However, it is important to use actual colon cancer tissue from patients having received chemotherapy and perform immunohistochemical staining, which can be easily used in a clinical setting.
CONCLUSIONS
Here, it has been shown that patients who responded to CPT-11, compared to those that did not, had a better prognosis. Also, the responsiveness to CPT-11 was shown to be higher in patients with loss of hMLH1 expression. Although, the numbers of the responder group were small, our results suggested that the hMLH1 protein expression status could be associated with CPT-11 responsiveness. This study indicates that the immunohistochemical determination of the MMR expression status may be valuable in selecting patients more likely to benefit from adjuvant chemotherapy with CPT-11. Further investigations in this field are warranted on the basis of these data.