AbstractPurposeThe purpose of this study was to investigate the concordance rate of PIK3CA mutations between primary and matched distant metastatic sites in patients with breast cancer and to verify whether there are differences in the frequency of PIK3CA hotspot mutations depending on the metastatic sites involved.
Materials and MethodsArchived formalin-fixed paraffin-embedded (FFPE) specimens of primary breast and matched distant metastatic tumors were retrospectively obtained for 49 patients. Additionally, 40 archived FFPE specimens were independently collected from different breast cancer metastatic sites, which were limited to three common sites: the liver, brain, and lung. PIK3CA mutations were analyzed using droplet digital polymerase chain reaction, including hotspots involving exons 9 and 20.
ResultsAfter analysis of 49 breast tumors with matched metastasis sites, 87.8% showed concordance in PIK3CA mutation status. According to PIK3CA hotspot mutation testing in 89 cases of breast cancer metastatic sites, the proportion of PIK3CA mutations at sites of metastasis involving the liver, brain, and lung was 37.5%, 28.6%, and 42.9%, respectively, which did not result in statistical significance.
ConclusionThe high concordance of PIK3CA mutation status between primary and matched metastasis sites suggests that metastatic sites, regardless of the metastatic organ, could be considered sample sources for PIK3CA mutation testing for improved therapeutic strategies in patients with metastatic breast cancer.
IntroductionBreast cancer is the most common malignancy in women. The World Health Organization indicates that there were 2.3 million women diagnosed with breast cancer globally in 2020. Most cases are diagnosed in the early stages; however, approximately 10%–41% of patients develop metastatic or advanced disease [1]. Hormone receptor (HR)–positive and human epidermal growth factor receptor 2 (HER2)–negative breast cancer account for 60%–70% of all breast tumors. The National Comprehensive Cancer Network (NCCN) guidelines recommend endocrine therapy with or without CDK4/6 inhibitors as a first-line treatment for post-menopausal HR+/HER2− advanced breast cancer [2].
Phosphoinositide 3 kinases (PI3Ks) are a family of lipid kinases that regulate the PI3K/AKT/mammalian target of rapamycin pathway involved in cell proliferation, adhesion, survival, and motility. PI3Ks are heterodimeric enzymes composed of catalytic and regulatory subunits and can be subdivided into three main classes (I–III) based on their structural, catalytic, and regulatory properties. Numerous studies have shown that PIK3CA, encoding the alpha isoform catalytic subunit of PI3K, is mutated in various cancers [3,4], resulting in constitutive enzymatic activity [5,6]. The PIK3CA mutation is the most frequent mutation identified in the HR+/HER2− subgroup among all breast cancers, with approximately 40% (range, 13% to 62%) of patients presenting with the mutation [7–9].
The initially developed PI3K inhibitors, pictilisib (GDC-0941, Genentech, Inc., South San Francisco, CA) and buparlisib (BKM120, Novartis, Basel, Switzerland), are pan-class I PI3K inhibitors. The addition of pictilisib to paclitaxel or fulvestrant does not significantly improve progression-free survival (PFS) in patients with advanced HR+/HER2− breast cancer. One potential reason is that dosing of the drug is limited by toxicity, which could limit its efficacy [10,11]. In the case of buparlisib, clinical trials have shown prolonged PFS compared with that of fulvestrant alone (BELLE-2, BELLE-3). The efficacy of buparlisib supports the rationale for using PI3K inhibitors in patients with PIK3CA mutations. However, frequent discontinuation due to adverse effects reduces the duration of treatment, potentially limiting the efficacy of such combination therapy [11–13]. The results of pan-class I PI3K inhibitors show the need for the development of PI3K inhibitors with greater selectivity in order to improve safety profiles and increase efficacy.
Isoform-selective PI3K inhibitors have been developed, and clinical studies have shown that such inhibitors exhibit reduced toxicity compared to pan-PI3K inhibitors [14]. The results of the SOLAR-1 Phase III trial provided evidence that the α-selective PIK3CA inhibitor alpelisib (BTL719, Novartis Pharma AG) combined with fulvestrant improved PFS in patients with HR+/HER2− PIK3CA-mutated advanced or metastatic breast cancer with a history of endocrine therapy [15]. Based on these results, the U.S. Food and Drug Administration (FDA) approved alpelisib in combination with fulvestrant for the treatment of post-menopausal patients diagnosed with HR+/HER2− PIK3CA-mutated advanced or metastatic breast cancer in May 2019. With the FDA approval of alpelisib, adequate testing for PIK3CA mutations to identify patients who are most likely to benefit from PI3K inhibitor therapy has become essential.
Considering the clinical benefit of PI3K inhibitor therapy in patients with HR+/HER2− PIK3CA-mutated advanced or metastatic breast cancer, determination of tumor PIK3CA mutational status is important for managing HR+/HER2− advanced or metastatic breast cancer. Whether the PIK3CA status of recent metastatic disease is more appropriate than that of the primary tumor is yet to be determined. Furthermore, not all tissue samples, whether primary or metastatic, are available for testing in clinical practice.
In this context, we investigated the concordance rate of PIK3CA mutations between primary breast tumors and matched metastases, and the difference in the proportion of PIK3CA somatic mutations involving three metastatic sites (the liver, brain, and lung) in breast cancer cases to provide information regarding appropriate specimen source sites for testing PIK3CA mutations in patients with metastatic breast cancer.
Materials and Methods1. Study designThe study protocol was approved by our Institutional Review Board (SMC IRB No. 2019-08-119). Archived formalin-fixed paraffin-embedded (FFPE) specimens of primary breast and matched remote metastatic sites were retrospectively obtained for 49 patients (study A). Subsequently, 40 additional archived FFPE specimens from breast cancer metastatic sites were independently acquired (study B). In studies A and B, the sites of metastasis were limited to three common organs in breast cancer: the liver, brain, and lung.
2. PathologyTumor histology and patient characteristics were extracted from pathology reports held at the Department of Pathology, Samsung Medical Center. Immunohistochemistry (IHC) for estrogen receptor (ER), progesterone receptor, human epidermal growth factor receptor 2 (HER2), and Ki-67 was performed separately for primary tumors and metastases. ER was considered positive if ≥ 1% of tumor cell nuclei exhibited staining [16]. HER2 was defined as being positive by IHC (3+) or by in situ hybridization if the HER2/chromosome 17 (CEP17) ratio was ≥ 2 and the number of HER2 gene copies was ≥ 4, or if HER2/CEP17 < 2 and HER2 copy number was ≥ 6, according to the American Society of Clinical Oncology–College of American Pathologists (ASCO-CAP) guidelines. Ki67 was assessed by IHC, and ≥ 20% positivity was considered high, as recommended by the St. Gallen consensus [17]. The intrinsic subtype of breast cancer was defined by a clinicopathological surrogate definition based on the 13th International Breast Cancer Conference held in St. Gallen, namely, luminal A, luminal B, HER2-enriched, and triple-negative breast cancer (TNBC) [17].
3. DNA extraction from FFPE tissuesFFPE tissue was sectioned as a 10 μm curl which was stored at room temperature. DNA was extracted from FFPE tissues using a Maxwell CSC DNA FFPE Kit (Promega Inc., Madison, WI) according to the manufacturer’s protocol. Extracted DNA was quantified using a Nanodrop instrument (Thermo Fisher Scientific Inc., Waltham, MA) and stored at −20°C in a freezer.
4. Droplet digital polymerase chain reaction process and analysisThe Droplex PIK3CA Mutation Test Kit (Gencurix Inc., Seoul, Korea) was used to quantitatively detect 18 PIK3CA mutation subtypes based on droplet digital polymerase chain reaction (ddPCR) technology. ddPCR primers and probe sets were designed to detect mutations involving PIK3CA: N345K, C420R, E542K, E545K, E545A, E545G, E545D, Q546K, Q546E, Q546R, E726K, M1043I, H1047R, H1047L, H1047Y, G1049R, and internal controls. Internal controls were used as validity indicators of the state of the specimen, DNA extraction, and PCR processes. All processes were performed according to the manufacturer’s protocols. Briefly, at least 45 ng/well (270 ng/6 wells) of DNA was used to detect PIK3CA mutations using ddPCR reagents (Supermix, Oligo Mix, Enzyme Mix, and DTT). Droplets were generated using a QX200 Droplet Generator (Bio-Rad, Hercules, CA) by loading 20 μL of the PCR amplification mix and 70 μL of Droplet Generation Oil for Probes (Bio-Rad) into each well of a DG8 cartridge (Bio-Rad). The droplet-oil mixture was transferred using an 8-channel electronic pipette to a semi-skirted 96-well plate (Bio-Rad). The plate was sealed with a pierceable foil heat seal using a PX1 PCR plate sealer (Bio-Rad). The 96-well plate was loaded onto a T100 Thermal Cycler (Bio-Rad) and run under the following thermal cycling conditions: 37°C for 30 minutes, 95°C for 10 minutes, followed by 40 cycles of 94°C for 30 seconds, 58°C for 1 minute, and 98°C for 10 minutes. After completion of the PCR process, the plate was read using a QX200 Droplet Reader with the following settings: channel 1, FAM; channel 2, HEX. After droplet reading, analysis was conducted using QuantaSoft software (Bio-Rad). The kit components contained uracil-DNA glycosylase to reduce deamination-induced nucleotide changes. The mutation index (MI) was calculated according to the protocol, and 1% of the MI of PIK3CA was adopted as the cutoff value in this study.
Results1. Baseline characteristics of patients with breast cancerEighty-nine Asian patients with metastatic breast cancer were included in this study. Archived FFPE blocks of primary breast and metastatic tumor sites (the liver, brain, and lung) were retrospectively collected to analyze PIK3CA mutations (Fig. 1). The clinicopathological characteristics of the 89 patients are presented in Table 1. The median age of the enrolled patients at the time of diagnosis of breast cancer was 51 years, with an age range of 31 to 82 years. Patients were classified as luminal A (n=25, 28.1%), luminal B (n=31, 34.8%), HER2 enriched (n=11, 12.4%), TNBC (n=9, 10.1%), and unknown (n=13, 14.6%) according to pathology reports of primary breast cancer tumors. The intrinsic subtype of breast cancer was mainly HR-positive (n=56, 62.9%), and PIK3CA mutations were significantly associated with HR expression (p=0.03). A total of 33 of 89 patients (37.1%) presented with PIK3CA mutations identified in remote metastatic malignancies.
2. Comparison of PIK3CA mutations and molecular subtypes between primary breast and matched metastatic tumorsTo analyze PIK3CA mutational stability, the concordance rate of PIK3CA mutations between primary tumors and matched distant metastases in patients with breast cancer was evaluated. In study A, 87.8% (43/49) showed concordance based on the results of the PIK3CA hotspot mutation test (Table 2). Calculating by using the PIK3CA primary breast cancer results as a reference, the positive percentage agreement (PPA), negative percentage agreement (NPA), and overall percentage agreement (OPA) were 85.0% (95% confidence interval [CI], 65.6 to 94.4), 89.7% (95% CI, 75.2 to 96.1), and 87.8% (95% CI, 75.2 to 95.4), respectively. Seven cases revealed discrepancies between the primary tumors and matched metastases, three of which had a mutation in PIK3CA exon 9 (E542K, n=1) or exon 20 (H1047R, n=2) only at the metastatic site, three patients had mutations in exon 20 (H1047R, n=2) or exon 7 (C420R, n=1) only in the primary breast tumor, and one patient harbored an additional mutation in exon 9 (E545D) at the metastatic site (Fig. 2A).
As shown in S1 Table, applying a cutoff value for the lower limit of detection (LOD) based on analytical validation of the kit changed the concordance rate of the PIK3CA mutation between breast tumors and matched metastases to 77.6% (38/49). PIK3CA mutation status was changed in a total of nine cases, of which seven had a low copy number of exon 9 (E542K or E545K) mutation in the primary breast tumor, and two had a low copy number of exon 20 (H1047R or M1043I) mutation at the metastatic sites.
In an analysis of molecular subtype alterations between primary breast malignancies and matched metastases, 15 patients (33.3%, 15/45) had altered molecular subtypes at the metastatic sites (Fig. 2B). Two patients without IHC staining results for metastases were excluded. The majority of subtype alterations were in luminal A (50.0%, 7/14), followed by luminal B (29.2%, 7/24) and TNBC (20.0%, 1/5). The results for each pair are described in S2 Table.
3. Frequency of PIK3CA mutations at sites of metastases and paired primary breast tumorsIn study B, 72.7% (24/33) of mutations were hotspot mutations involving the kinase domain (exon 20), 18.2% (6/33) were hotspot mutations in the helical domain (exon 9), and 9.1% (3/33) of mutations were found in other domains, such as exons 4 or 13, in metastatic sites of breast cancer (Fig. 3A). Multiple PIK3CA mutations were found in three patients, two of whom had the E726K mutation (Fig. 3B).
In study A, the frequency of PIK3CA mutations was 45.8%, 30.0%, and 40.0% in primary breast cancer, which showed liver, brain, and lung metastases, respectively. The frequencies of PIK3CA mutations in the matched liver, brain, and lung metastases were 50.0%, 30.0%, and 33.3%, respectively (Table 3). To evaluate the prevalence of PIK3CA mutations among the sites of metastases, metastatic lesions were classified as liver and others, brain and others, and lung and others. Based on the results of the PIK3CA mutation test in primary breast tumors, differences in mutation frequencies in liver, brain, and lung metastases did not attain statistical significance (p=0.68, p=0.50, and p > 0.99, respectively). In study B, the proportions of PIK3CA mutations in the liver, brain, and lungs were 37.5%, 28.6%, and 42.9%, respectively (Table 3). In brain metastases, PIK3CA mutations showed a lower frequency than in that in liver and lung metastases; however, there was no significant difference in PIK3CA mutation frequencies between the brain and other metastases. As shown in Fig. 4, the prevalence of PIK3CA mutations among the metastatic sites was not significantly different. In the sub-analysis based on HR expression, there was no significant difference in mutation frequency according to the metastatic site (S3 Fig.).
DiscussionBased on a randomized phase III clinical trial of patients with HR+ and PIK3CA-mutated advanced breast cancer [18], PIK3CA mutations have reached category 1 of the NCCN category of evidence [2], which indicates a uniform NCCN consensus for clinical intervention. Therefore, collecting appropriate samples for testing for PIK3CA mutations is important in identifying patients who are most likely to benefit from treatment with PI3K inhibitors. In this context, we investigated (1) the concordance rate of PIK3CA mutations between primary breast cancer and matched metastases and (2) the frequency of PIK3CA somatic mutations, depending on the sites of metastatic breast cancer.
Previous studies have shown that PIK3CA is one of the most frequently mutated genes in human breast cancer [7,19,20]. The PIK3CA mutational frequency differed according to molecular subtype, with a high frequency in HR-positive breast cancer cases and a low frequency in TNBC [19]. Similar to previous studies, 26 cases (46.4%) of HR+ breast cancers showed PIK3CA mutations at breast cancer metastatic sites (Table 1).
In May 2019, the FDA-approved alpelisib in combination with fulvestrant for post-menopausal women and men with HR-positive, HER2-negative, PIK3CA-mutant advanced/metastatic breast cancer following progression on or after an endocrine-based therapeutic regimen. There are three FDA-approved companion diagnostics for alpelisib to select patients with PIK3CA mutations: Therascreen PIK3CA RGQ PCR Kit (Qiagen, Hilden, Germany), FoundationOne CDx, and FoundationOne Liquid CDx (Foundation Medicine, Inc., Cambridge, MA). As described in the kit indications, the Therascreen PIK3CA RGQ PCR kit uses genomic DNA extracted from FFPE breast tumor tissues or circulating tumor DNA (ctDNA) from K2EDTA plasma. The contingency table shows PPA, NPA, and OPA between PIK3CA ctDNA and PIK3CA tissue results are reported to be 54.6%, 97.2%, and 71.5%, respectively [21]. Based on the contingency table, the FDA recommends reflex testing using tissue specimens if ctDNA test results are negative.
According to the concordance rate of PIK3CA mutations between primary and matched metastatic sites, the latter could be considered as specimens for PIK3CA mutation screening for therapeutic strategies in patients with breast cancer. As shown in Table 2, the concordance rate of PIK3CA hotspot mutations was 87.8% (43/49) between matched primary breast tumors and metastases, indicating that PIK3CA hotspot mutations are highly stable between primary breast tumors and matched metastases. The PPA, NPA, and OPA between PIK3CA screening results for metastatic sites and primary sites were 85.0%, 89.7%, and 87.8%, respectively, showing a much higher PPA than that from PIK3CA ctDNA results [21]. Considering the high PPA values, metastatic sites could be considered as sampling sources for PIK3CA mutation testing. In the case of changes in molecular subtype, only 66.7% (30/45) maintained the subtype on metastatic sites involving the lung, brain, or liver when analyzed except for two unknown cases (Fig. 2, S2 Table).
Although ddPCR assays can precisely quantify target DNA at high levels of sensitivity and specificity [22–24], as companion diagnostics, evaluating appropriate cutoff values considering clinical utility is important in order to develop molecular diagnostic assays in clinical settings. We analyzed ddPCR data using a cutoff MI value of 1%, which is a threshold above the lower LOD of the kit. MI refers to the percentage of mutant to total PIK3CA in DNA extracted from FFPE blocks [25]. To evaluate changes in the concordance rate of PIK3CA mutations between breast tumors and matched metastases, we applied a cutoff value of the lower LOD based on analytical validation of the kit (S1 Table). The concordance rate decreased to 77.6% (38/49). This might be a result of the higher sensitivity of the ddPCR method, but considering that the samples were immobilized in FFPE blocks, artifacts related to formalin deamination may be present and falsely affect the concordance rate between primary breast malignancies and matched metastases [26,27]. Seven of nine cases with altered mutation detection results based on cutoff values of the lower LOD were in exon 9, 1624 G>A (E542K) or 1633 G>A (E545K) substitution mutation, which is associated with formalin-induced deamination. Care must be taken when interpreting data analyzed from FFPE tissues.
PIK3CA mutations at the site of metastasis were mainly detected in the kinase domain (exon 20, 72.7%) and helical domain (exon 9, 18.2%) (Fig. 3A), and multiple PIK3CA mutations were found in three (9.1%) cases, and two of these harbored the E726K mutation (Fig. 3B). In 2019, a study reported that 12% of PIK3CA mutations involve multiple mutations in breast cancer, and E726, E453, and M1043 mutations are major mutations enriched in multiple tumors in breast cancer datasets. Moreover, increased sensitivity to PI3K inhibitors is observed in double PIK3CA mutations compared to that in single hotspot mutations [28]. Although we analyzed PIK3CA hotspot mutations involving metastatic sites of breast cancer, the mutation frequencies were similar to those reported in previous studies [8,28,29]. Clear identification of PIK3CA mutation profiles is essential for the effective use of therapeutics. As data concerning PIK3CA mutation profiles associated with therapeutic responses accumulate, it is expected that the clinical performance of PI3K inhibitors will be improved.
A number of studies have reported a significant difference in the PIK3CA mutation rate depending on the site of metastasis [30]. However, according to the present study, there was no statistically significant difference in the frequency of PIK3CA mutations among liver, brain, and lung metastases of patients with breast cancer (Fig. 4). A sub-analysis based on HR expression also showed no statistically significant differences between metastatic sites (S3 Fig.), particularly in HR+ breast cancers. Only three cases of brain metastases involved PIK3CA mutations at the site of metastases in HR-negative breast cancer, but there were no statistically significant differences (p=0.22) in the frequency of PIK3CA mutations among liver, brain, and lung metastases. In HR-negative breast cancer, the relatively small sample size (n=20) might not have had sufficient statistical power to reveal statistical significance. Further studies are needed to evaluate differences in PIK3CA mutation frequencies between metastatic sites in HR-negative breast cancer. After metastasis, PIK3CA mutations remained stable, regardless of metastatic site. Comparison of PIK3CA mutation rates in primary breast and matched metastatic tumors according to the metastasis site showed highly consistent results (Table 3).
Our study has certain limitations. First, archived FFPE samples were retrospectively collected, which could cause selection bias. Additionally, only patients with liver, brain, and lung metastases were included, although these are common sites of metastasis in breast cancer cases. Despite these limitations, the high concordance of PIK3CA mutation status between primary tumors and matched metastases reported in the present study suggested that metastatic sites, regardless of the metastatic organ, could be considered as specimens for testing PIK3CA mutations for therapeutic strategies in patients with metastatic breast cancer. If a metastatic carcinoma sample cannot be obtained, PIK3CA mutation testing may also be performed on the primary tumor sample.
Electronic Supplementary MaterialSupplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
NotesEthical Statement The study protocol was approved by our Institutional Review Board (SMC IRB No. 2019-08-119). A written informed consent was obtained from all study participants. Author Contributions Conceived and designed the analysis: Choi YL, Park J. Collected data: Sung M, Park J. Contributed data or analysis tools: Park J, Chang ES, Song JY, Jung K. Performed analyses: Park J, Cho SY, Kim SS. Wrote the paper: Park J. Supervision and revision of the manuscript: Cho SY, Choi YL, Shin YK. AcknowledgementsThis research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare (HI19C0141), the BK21FOUR Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education (5120200513755), and NRF grants funded by the Ministry of Science, ICT and Future Planning (2016R1A5A2945889 and 2019R1A2B5B02069979), Republic of Korea.
Fig. 2Alteration of PIK3CA mutations (A) and molecular subtype classifications (B) between primary breast cancer and matched metastases in 49 patients with breast cancer. HER2, human epidermal growth factor receptor 2; ND, not detected; TNBC, triple-negative breast cancer. 
Fig. 3Mutations involving PIK3CA in metastatic sites of breast cancer. (A) Frequency of PIK3CA mutation in 33 patients who had PIK3CA mutation in the site of metastasis. In the case of multiple PIK3CA mutations, only the major mutation based on the mutation index (MI) was included. (B) Cases of multiple mutations in PIK3CA. The classification of major and minor mutations was based on MI (%). ABD, adaptor binding domain; RBD, ras-binding domain. 
Fig. 4
PIK3CA mutation proportions of 89 patients with metastatic breast cancer by site of metastasis. Mosaic plots representing PIK3CA mutation proportion for the 89 patients of metastatic breast cancer by site of metastases. PIK3CA mutation screening was performed at the site of metastases using droplet digital polymerase chain reaction assays. p-values were calculated using chi-square tests. 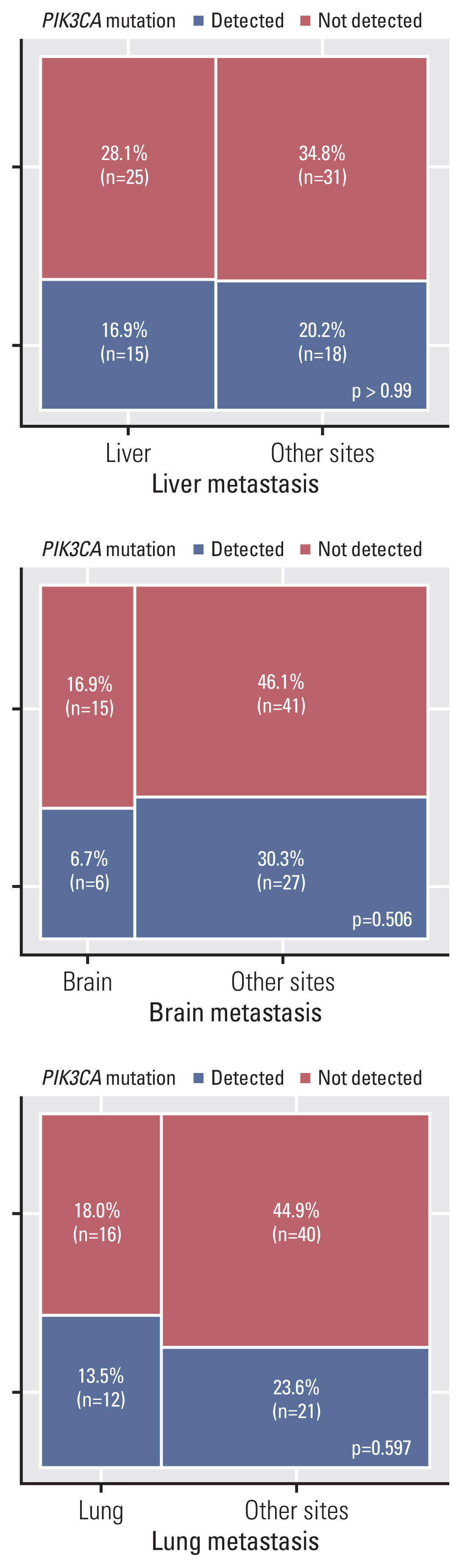
Table 1Baseline characteristics of patients with breast cancer (n=89) Table 2Concordance rate for PIK3CA mutations between primary and matched metastatic sites
Table 3The proportion of PIK3CA mutations in study A and study B References1. Pan H, Gray R, Braybrooke J, Davies C, Taylor C, McGale P, et al. 20-Year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N Engl J Med. 2017;377:1836–46.
2. Gradishar WJ, Anderson BO, Abraham J, Aft R, Agnese D, Allison KH, et al. Breast cancer, version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2020;18:452–78.
3. Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol. 2018;15:273–91.
4. Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94:455–9.
5. Ligresti G, Militello L, Steelman LS, Cavallaro A, Basile F, Nicoletti F, et al. PIK3CA mutations in human solid tumors: role in sensitivity to various therapeutic approaches. Cell Cycle. 2009;8:1352–8.
6. Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–1000.
7. Mosele F, Stefanovska B, Lusque A, Tran Dien A, Garberis I, Droin N, et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol. 2020;31:377–86.
8. Anderson EJ, Mollon LE, Dean JL, Warholak TL, Aizer A, Platt EA, et al. A systematic review of the prevalence and diagnostic workup of PIK3CA mutations in HR+/HER2− metastatic breast cancer. Int J Breast Cancer. 2020;2020:3759179.
9. Deng L, Zhu X, Sun Y, Wang J, Zhong X, Li J, et al. Prevalence and prognostic role of PIK3CA/AKT1 mutations in Chinese breast cancer patients. Cancer Res Treat. 2019;51:128–40.
10. Vuylsteke P, Huizing M, Petrakova K, Roylance R, Laing R, Chan S, et al. Pictilisib PI3Kinase inhibitor (a phosphatidylinositol 3-kinase [PI3K] inhibitor) plus paclitaxel for the treatment of hormone receptor-positive, HER2-negative, locally recurrent, or metastatic breast cancer: interim analysis of the multicentre, placebo-controlled, phase II randomised PEGGY study. Ann Oncol. 2016;27:2059–66.
11. Krop IE, Mayer IA, Ganju V, Dickler M, Johnston S, Morales S, et al. Pictilisib for oestrogen receptor-positive, aromatase inhibitor-resistant, advanced or metastatic breast cancer (FERGI): a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2016;17:811–21.
12. Baselga J, Im SA, Iwata H, Cortes J, De Laurentiis M, Jiang Z, et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:904–16.
13. Di Leo A, Johnston S, Lee KS, Ciruelos E, Lonning PE, Janni W, et al. Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor-positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2018;19:87–100.
14. Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer. 2019;18:26.
15. Andre F, Ciruelos EM, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib (ALP)+fulvestrant (FUL) for advanced breast cancer (ABC): results of the phase III SOLAR-1 trial. Ann Oncol. 2018;29(Suppl 8):VIII709.
16. Allison KH, Hammond MEH, Dowsett M, McKernin SE, Cary LA, Fitzgibbons PL, et al. Estrogen and progesterone receptor testing in breast cancer: ASCO/CAP guideline update. J Clin Oncol. 2020;38:1346–66.
17. Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–23.
18. Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380:1929–40.
19. Cizkova M, Susini A, Vacher S, Cizeron-Clairac G, Andrieu C, Driouch K, et al. PIK3CA mutation impact on survival in breast cancer patients and in ERalpha, PR and ERBB2-based subgroups. Breast Cancer Res. 2012;14:R28.
20. Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–81.
21. U.S. Food and Drug AdministrationSummary of safety and effectiveness data of therascreen PIK3CA RGQ PCR kit [Internet]. Silver Spring MD: U.S. Food and Drug Administration; 2019. [cited 2022 Jan 2]. Available from: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P190004
22. Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y, et al. Highly sensitive droplet digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. J Mol Diagn. 2015;17:265–72.
23. Olmedillas-Lopez S, Garcia-Arranz M, Garcia-Olmo D. Current and emerging applications of droplet digital PCR in oncology. Mol Diagn Ther. 2017;21:493–510.
24. Taylor SC, Laperriere G, Germain H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci Rep. 2017;7:2409.
25. Kim SS, Choi HJ, Kim JJ, Kim MS, Lee IS, Byun B, et al. Droplet digital PCR-based EGFR mutation detection with an internal quality control index to determine the quality of DNA. Sci Rep. 2018;8:543.
26. Kim S, Park C, Ji Y, Kim DG, Bae H, van Vrancken M, et al. Deamination effects in formalin-fixed, paraffin-embedded tissue samples in the era of precision medicine. J Mol Diagn. 2017;19:137–46.
27. Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61:64–71.
28. Vasan N, Razavi P, Johnson JL, Shao H, Shah H, Antoine A, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kalpha inhibitors. Science. 2019;366:714–23.
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||