AbstractPurposeFor the target treatment and prevention of women’s increased thyroid cancer, we focused on risks of environmental exposure to endocrine disrupting chemicals, particularly bisphenol A (BPA), and its high susceptible exposure-timing, particularly early exposure in lives.
Materials and MethodsFemale ICR mice were exposed to BPA in utero and in early life (15, 75, and 300 mg/L of drinking water via pregnant mice and lactation). We identified BPA-responsive proteins in mice thyroid by two-dimensional gel electrophoresis, image analyses, and electrospray ionization quadrupole time-of-flight mass spectrometry. We further analyzed expression of the BPA-responsive proteins in women thyroid cancer patients (n=28).
ResultsWe found the altered 17 proteins in BPA dose-dependent manner among the thyroid tissues of offspring mice and identified nine proteins of them, including Anxa6, Atp5b, Hspa5, and Vcp, etc. In addition, we observed the positive association between blood BPA levels and mRNA expression of the ANXA6 and VCP not in normal but thyroid cancer tissues.
IntroductionBisphenol A [4,4’-(propane-2, 2-diyl)diphenol, BPA] has been speculated as an endocrine disrupting chemical (EDC); however, people are easily exposed to BPA in their daily lives, because of its wide use as glass substitutes [1]. BPA is detected in urine, blood, placenta, breast milk, and even umbilical cord [2]. Numerous studies also suggest that BPA perturbs thyroid functions through multiple mechanisms. For example, a recent Zebrafish study showed BPA interferences with thyroid specific gene expression [3]. In addition, BPA exposure seems to elevate triiodothyronine, while decrease thyroxin and thyroid-stimulating hormone (TSH) [4]. Therefore, thyroid can be a target organ of BPA toxicity [5]. We recently reported BPA is metabolized into toxic metabolites by cytochrome P450 2E1 [1]; however, its consistent mechanisms between in vitro or in vivo and human are not clear, yet.
Among thyroid diseases, thyroid cancer has been emphasized due to its incidence rate continuously increased across the world over the last three decades [6]. The dramatic increase in thyroid cancer incidences might be related to the advances of diagnostic technologies, overdiagnosis [7], environmental and lifestyle factors, etc. [8]. However, the increased medical surveillance or improved detection methods alone cannot fully explain the increase. Multiple factors might be associated together for risks of thyroid cancer [9]. Particularly, hormonal and reproductive factors have shown significant associations with thyroid cancer risks [10]. The estrogens have been conjectured to be one of the main causes of thyroid diseases [11] because of the higher thyroid cancer rate and an earlier peak rate of women than men. Therefore, environmental estrogens, i.e., EDCs, have been suspected as risk factors, but little is known about their risks for human thyroid cancer.
A few studies have explored the aspect of toxic end points of BPA. The results suggested the theory of ‘two hits’ or ‘second hit’ for instance hormone and BPA exposure as an alternative mechanism of BPA mutagenesis or carcinogenesis [12,13]. In addition, some studies suggested epigenetic alterations or genetic memory as BPA toxic mechanisms [14]. Particularly, the Developmental Origins of Health and Disease (DOHaD) hypothesis can be applied to the consequences of epigenetic memory or reprogramming. In other words, environmental and nutritional factors influence developmental plasticity during the critical periods of development, thereby altering susceptibility to diseases later in life [15]. Therefore, many researchers have highlighted the importance of exposure timing [16]. Therefore, we focused on risks of environmental exposure to BPA and its high susceptible exposure-timing, particularly early exposure in lives, for the target treatment and prevention of women’s increased thyroid cancer. We designed in utero and early life exposure to BPA in mice and identified BPA-responsive protein biomarkers in thyroid tissues of female offspring mice. We also confirmed these BPA exposure biomarkers in women thyroid cancer patients.
Materials and Methods1. Animal care and treatmentICR mice in the first week of pregnancy were obtained from Orient Co. (Seoul, Korea) and housed under a 12-hr light/dark cycle (lights on 07:00-19:00) at 21-24°C and 40%-60% humidity with unlimited access to drinking water. The pregnant mice were fed with pellet food (Samtako, Seoul, Korea) ad libitum, and were administered three different concentrations of BPA (Sigma-Aldrich Co., St. Louis, MO), 15, 75, and 300 mg/L dissolved in 0.1% ethanol/water using glass bottles for 34-36 days, i.e., from day 7 of pregnancy to lactation (day 21 post-delivery). After the lactation, the offspring mice were allowed an access to untreated water. Fifteen or sixteen pregnant mice were administered with each concentration of BPA. In contrast, untreated control pregnant mice (n=18) were provided with 0.1% ethanol/water ad libitum. The 7-week-old female offspring were sacrificed in CO2 chamber and obtained their thyroids. All animal experimental procedures were complied with the University Animal Care and Use Committee at Sookmyung Women’s University.
2. Proteomic analysesTotal protein was extracted from 10 mg of the organs using our previous method [17] with minor modification. After centrifuging the homogenized samples (14,000 rpm for 10 minutes), we measured the protein concentrations using the Bradford assay. Two-dimensional gel electrophoresis (2D-PAGE) and image analyses were performed with the extracted 300 μg of each protein sample. The digitalized image was analyzed with PDQUEST software (ver. 6.1, Bio-Rad, Richmond, CA). After analysis of the nano-electrospray ionization on a quadrupole time-of-flight (Q-TOF) mass spectrometer (Micromass, Manchester, UK) with 30 μL of digested peptide mixture, all tandem mass spectrometry (MS/MS) spectra derived from spot were identified by NCBInr and EST databases using the MASCOT search program [17]. The optical density of specific proteins was statistically analyzed with the Mann-Whitney test (significance at p < 0.05). We duplicated the 2D gel analyses with two different mice for each treatment (control, 15, 75, and 300 mg BPA/L of water, total=8).
3. Thyroid tissues from cancer patientsMatched pairs of thyroid tumor and adjacent non-tumor tissues were collected from women thyroid cancer patients (n=28; age, 47.4±11.6 years; papillary carcinoma, 100%; metasatasis, 10.4% of the subjects), who received a surgical treatment at Hanyang University Hospital, Seoul, Korea. Their tissues were stored in sterile e-tubes and frozen at –80°C until RNA isolation. All of the procedures for the human study were approved by the Clinical Research Ethics Committee of Hanyang University Hospital.
4. Real-time polymerase chain reaction analysisTotal RNA was isolated from thyroid tumor and normal tissues in each patient (~20 mg) with Trizol reagent (Thermo Fisher Scientific, Carlsbad, CA) according to the manufacturer’s instructions. The RNA quality was assessed by conventional agarose gel electrophoresis, and RNA concentration was determined using a Qubit fluorometer (Thermo Fisher Scientific). Total RNA (1 μg) from each sample was reverse transcribed to cDNA using a TaqMan RT Reagent kit (Thermo Fisher Scientific). We performed quantitative real-time polymerase chain reaction (PCR) analyses with TaqMan Gene Expression Assays [17]. Taqman qPCR primers and probes were as follows: ATP5B (Hs00969569_m1), ANXA6 (Hs00241762_m1), HSPA5 (Hs00607129_gH), and valosin-containing protein (VCP; Hs00997642_m1). The reaction was run on a 7500 Real-Time PCR System (Thermo Fisher Scientific) under the following conditions: 2 minutes at 50°C for AmpErase UNG activation and 10 minutes at 95°C for UNG inactivation, followed by 40 cycles of 15 seconds at 95°C for denaturing, and 1 minute at 60°C for annealing and extension. After constructing standard curves, we quantified mRNA expression levels of ATP5B, ANXA6, HSPA5, and VCP, and normalized with respect to the expression of 18S rRNA, of which primers and probes were obtained from Thermo Fisher Scientific.
5. Analyses of blood BPA in thyroid cancer patientsBPA levels in blood specimens were analyzed using reverse phase-HPLC/FLD via liquid-liquid extraction with minor modifications [2]. In brief, 500 μLof blood sample and 50 μL of 4.13 uM bisphenol B (internal standard) were mixed with 30 μL of 2.0 M sodium acetate (pH 5.0), and hydrolyzed with 20 μL of β-glucuronidase (2,000 U) for 5 hours at 37°C in a shaking water bath. After hydrolysis, 100 μL of 2.0 M HCl was added and the mixtures were extracted twice with 5 mL of ethylacetate. Six milliliters of the supernatant was evaporated in a Savant SpeedVac centrifuge (Savant Instruments Inc., Holbrook, NY) and dissolved in 300 μL of 60% acetonitrile. One hundred microliters of the resultant was injected into the HPLC/FLD apparatus. The HPLC/FLD system was the same with the previous one [2]. Samples were analyzed with gradient elution with water (solvent A) and acetonitrile (solvent B) (ratio of A to B, 0-35 minutes, 75:25 to 52:48; 35-40 minutes, 52:48 to 0:100; 40-45 minutes, 0:100-0:100; 45-55 minutes, 0:100 to 75:25; 55-70 minutes, 75:25). The samples were analyzed at 225 nm (excitation) and 305 nm (emission) with 1 mL/min of flow.
6. Statistical analysisShapio-Wilk W test was used to examine the degree of normal distribution of BPA levels. The normality of the expression data was verified using the Kolmogorov-Smirnor test. The Wilcoxon signed rank test was used to compare levels of gene expression of ANXA6, ATP5B, HSPA5, and VCP between tumor and adjacent non-tumor tissues. Pearson’s correlation analysis was used to determine whether there were differences in correlation of the gene expressions between tumor and non-tumor tissues. All statistical analyses were conducted using the JMP ver. 4.0.2 package (SAS Institute Inc., Cary, NC).
ResultsWhen the pregnant ICR mice were administered 15, 75, and 300 mg of BPA/L in water from day 7 of pregnancy to lactation, the BPA intake for each group was estimated at approximately 8.9±1.8 mg/kg/day, 47.1±5.8 mg/kg/day, and 171.1 ±16.8 mg/kg/day, respectively. These levels were established by the lowest-observed adverse-effect level (50 mg/kg/day) of BPA [18]. There were no statistically significant differences in offspring’s body weight due to BPA exposure.
Fig. 1 shows a representative 2D-PAGE gel of the proteomic analysis. Comparison of abundant proteins on 2D gels using the PDQUEST program revealed that the expression of 17 proteins was significantly changed in a dose-dependent manner (Fig. 1): 11 proteins were upregulated, and six downregulated by BPA exposure. However, we identified nine proteins out of the 17 proteins using the electrospray ionization Q-TOF mass spectrometry analysis (Table 1).
Women’s resected thyroid tumor and adjacent nontumor tissues were used to compare the expression of BPA-responsive proteomic biomarkers. The protein biomarkers identified in mice were examined for human. Four genes, ANXA6, ATP5B, HSPA5, and VCP, were selected based on their molecular functions as potential BPA-responsive markers in human. Three genes, ANXA6, HSPA5, and VCP, were up-regulated by BPA in mice thyroid tissues (Table 1). Expression of the four genes was assessed using matched tumor and adjacent nontumor tissues (n=28). There were no significant differences in the gene expression between the tumor and adjacent nontumor tissues (p > 0.05) (Fig. 2), although the mean expression level of VCP was somewhat higher in the tumor tissues than the adjacent non-tumor tissues.
BPA was detected in 90% of patients’ blood with the HPLC/FLD system. The limit of quantification was 0.39 ng/mL. The range of total BPA in the blood was 0-10.32 ng/mL (Fig. 3). The validity of BPA protein biomarkers from mice was examined in thyroid cancer patients by correlating BPA levels and gene expression. The results revealed strong associations between blood BPA levels and expression of the VCP or ANXA6 in thyroid tumor tissues (Fig. 4). In contrast, this trend was not shown in thyroid normal tissues.
DiscussionBPA showed proteomic alteration in various organs. For example, BPA negatively affected sperm motility, viability, mitochondrial functions, and intracellular ATP levels by activating the mitogen-activated protein kinase, phosphatidylinositol 3-kinase, and protein kinase-A pathways and induced differential expressions of 24 proteins in vitro [20]. We also reported that some of BPA-responsive proteins, Apo-AI, DppIII, Set and Vat, were involved in the anti-inflammatory response and tumorigenesis in immune organs, e.g., the thymus and spleen of mice [17,21]. In addition, some in vivo studies showed the impact of BPA on the thyroid. For example, BPA exposure in pregnant mice induced CpG hypomethylation of the promoter region of Cdk5 activator-binding protein (CabpIAP) [22] and Cdk5 activity via Rb is known to be critical to tumorigenesis and progression of medullary thyroid carcinoma [23]. However, these findings are not confirmed in human, yet. Therefore, the present study was performed to fill the gap between humans and animals and was focused on how BPA interferes thyroid cancer via BPA-responsive proteins.
We identified the BPA-responsive proteins in the thyroids of pregnant mice (Table 1). Among these, nine proteins are involved in the regulation of lipid metabolism, protein catabolism, and ubiquitin/proteasome-degradation pathways. We further corroborated these findings by using thyroid tumor tissues from women thyroid cancer patients to determine whether BPA exposure affects thyroid cancer. Our study revealed a significant correlation between the levels of BPA in blood and the gene expression of the BPA responsive proteins, e.g., ANXA6 and VCP, in thyroid tissues. Annexin A6 belongs to the highly conserved annexin protein family. The function of ANXA6 is linked to its ability to bind phospholipids in a Ca2+-dependent manner, thereby interacting with cellular membranes in a dynamic, reversible, and regulated fashion. It is also considered to be a tumor suppressor that is downregulated in squamous cervical cancer [24], breast cancer [25] via RAS/MAPK or EGFR signaling pathways. ANXA6 also increased anchorage-independent growth in breast cancer cells [26]. In the thyroid gland, TSH modulated ATP-induced Ca2+ signaling via ANXA6 in thyrocytes [27]. Similarly, Xiong et al. [28] suggested that ANXA6 is a potential biomarker for papillary thyroid carcinoma with lymph node metastasis via proteomic analyses in patients. Therefore, our results suggest an involvement of BPA in thyroid carcinogenesis via ANXA6.
The protein levels of VCP, an inflammation regulator, also called various cellular activities (AAA+) chaperone p97, were up-regulated in mice and its mRNA expression in women’s thyroid tumor tissues (Fig. 4). VCP is known to govern critical steps in ubiquitin-dependent protein quality control and intracellular signaling pathways [29]. In addition, VCP expression was correlated with lymph node metastasis and low survival rate in thyroid cancer [30]. Therefore, the up-regulation of Vcp by BPA (Table 1) suggests that thyroid function may be disrupted via VCP-mediated inflammatory pathways. However, there were no statistically significant differences between non-tumor and tumor tissues in the expression levels of the ANXA6 or VCP in the present study. Therefore, BPA may accelerate tumor progress rather than tumor incidence via ANXA6 or VCP. Our previous study showed that BPA is not mutagenic but stimulates MNNG mutagenicity [13]. In addition, the second hit theory or two-hit hypothesis can explain BPA-involvement in thyroid cancer via epigenetic memory with clear carcinogens [12,13].
In the near future, we will confirm the present results in enlarged number of human subjects to overcome the study limitation, particularly small size of the human tissues, and to obtain reproducibility and reliability for the above BPA-responsive biomarkers.
In conclusion, our study provides ANXA6 and VCP as proteomic biomarkers for BPA–early life exposure and their potential for women’s thyroid cancer.
AcknowledgmentsThis work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIP) (No. 2011-0030074).
Fig. 1.The 2D gel image shows 17 protein spots were altered in bisphenol A (BPA) dose-dependent manner on thyroid tissue of female offspring by prenatal exposure to BPA. 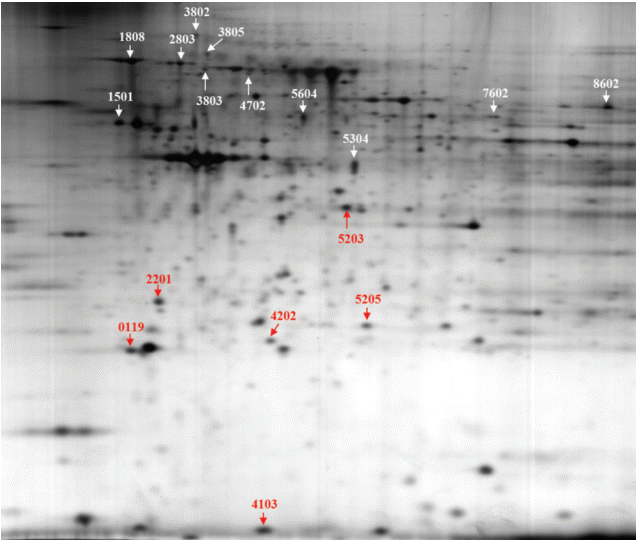
Fig. 2.No differences in expression of ANXA6, ATP5B, HSP5, or VCP between normal and tumor thyroid tissues in the women thyroid patients. N, normal; T, tumor. 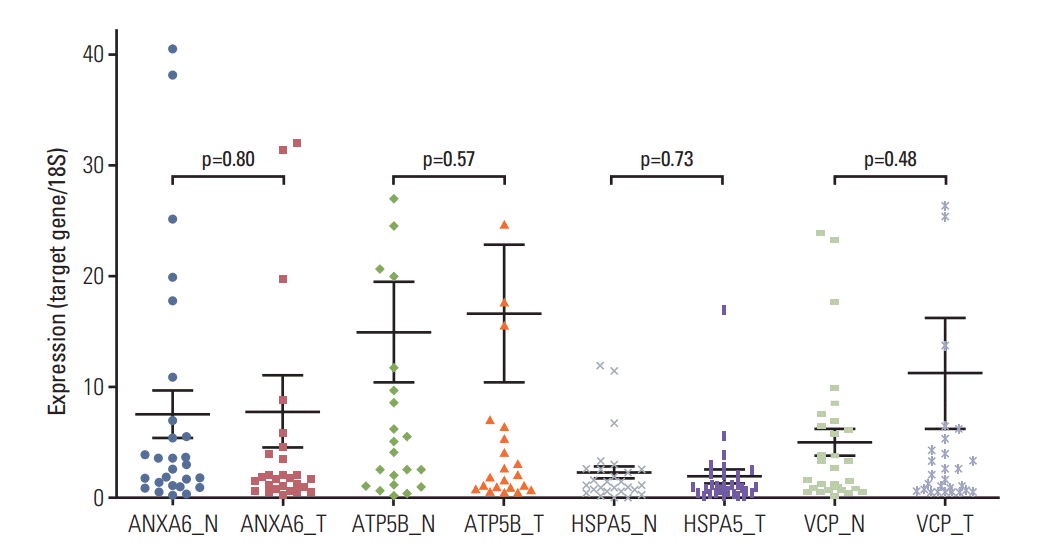
Fig. 3.Distribution of blood bisphenol A (BPA) levels. Upper part of the figure shows an outlier box plot with the square in the box showing the interquartile range: median, 1.38 μg/L. 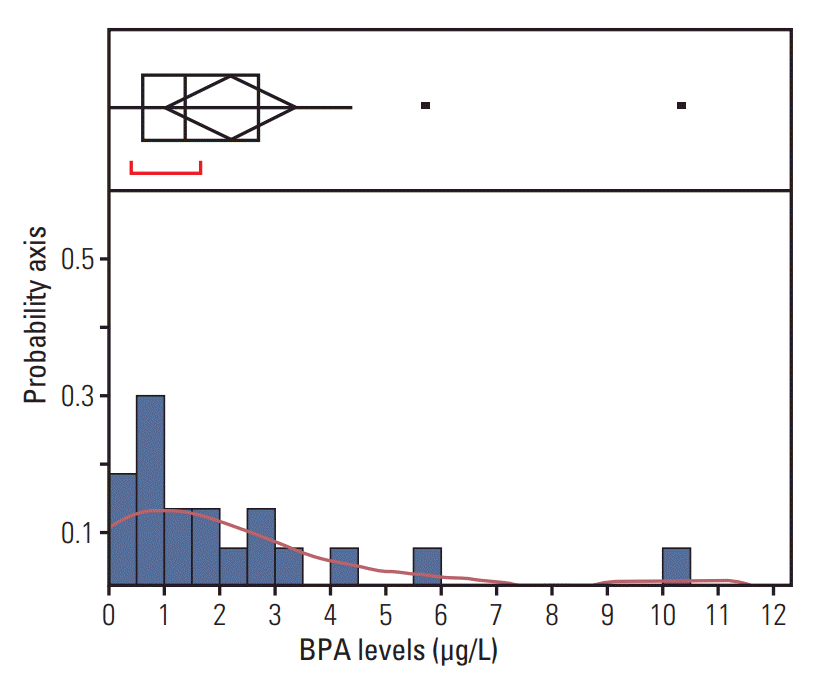
Fig. 4.The positive associations between blood bisphenol A (BPA) levels and gene expressions of ANXA6 or VCP in tumor rather than normal tissues among the women thyroid cancer patients. Upper blue line, VCP in tumor tissues; lower blue line, ANXA6 in tumor tissues; upper red line, VCP in normal tissue; lower red line, ANXA6 in normal tissues; N, normal; T, tumor; For the view of figure, we excluded an outlier, who had 10.32 μg/L of blood BPA by outlier test of Grubbs [19]. 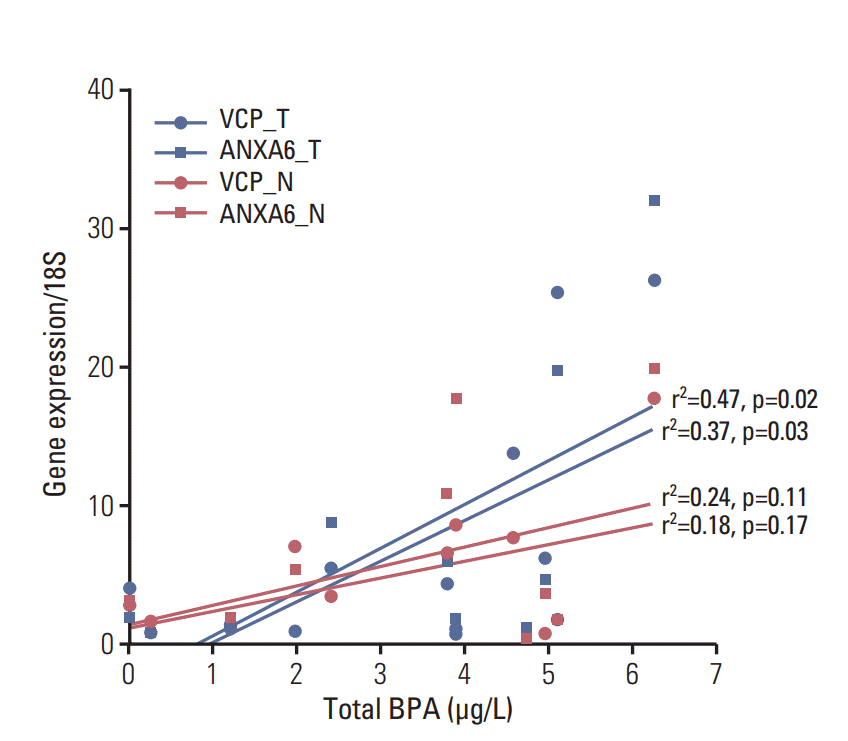
Table 1.Identification of differentially expressed proteins by BPA-dose dependent manner in mice
a) Location of the protein resolved by two-dimensional gel electrophoresis (Fig. 1) and identified by matrix assisted laser desorption ionization–time of flight mass spectrometry, References1. Lee DW, Oh WY, Yi SH, Ku B, Lee MY, Cho YH, et al. Estimation of bisphenol A: human toxicity by 3D cell culture arrays, high throughput alternatives to animal tests. Toxicol Lett. 2016;259:87–94.
2. Yi B, Kim C, Yang M. Biological monitoring of bisphenol A with HLPC/FLD and LC/MS/MS assays. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:2606–10.
3. Ghotbaddini M, Powell JB. The AhR ligand, TCDD, regulates androgen receptor activity differently in androgen-sensitive versus castration-resistant human prostate cancer cells. Int J Environ Res Public Health. 2015;12:7506–18.
4. Andrianou XD, Gangler S, Piciu A, Charisiadis P, Zira C, Aristidou K, et al. Human exposures to bisphenol A, bisphenol F and chlorinated bisphenol A derivatives and thyroid function. PLoS One. 2016;11:e0155237
5. Andra SS, Makris KC. Thyroid disrupting chemicals in plastic additives and thyroid health. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2012;30:107–51.
6. Yu GP, Li JC, Branovan D, McCormick S, Schantz SP. Thyroid cancer incidence and survival in the national cancer institute surveillance, epidemiology, and end results race/ethnicity groups. Thyroid. 2010;20:465–73.
7. Lee S, Lee YY, Yoon HJ, Choi E, Suh M, Park B, et al. Responses to overdiagnosis in thyroid cancer screening among Korean women. Cancer Res Treat. 2016;48:883–91.
9. Pellegriti G, Frasca F, Regalbuto C, Squatrito S, Vigneri R. Worldwide increasing incidence of thyroid cancer: update on epidemiology and risk factors. J Cancer Epidemiol. 2013;2013:965212.
10. Caini S, Gibelli B, Palli D, Saieva C, Ruscica M, Gandini S. Menstrual and reproductive history and use of exogenous sex hormones and risk of thyroid cancer among women: a meta-analysis of prospective studies. Cancer Causes Control. 2015;26:511–8.
11. Oh CM, Jung KW, Won YJ, Shin A, Kong HJ, Lee JS. Age-period-cohort analysis of thyroid cancer incidence in Korea. Cancer Res Treat. 2015;47:362–9.
12. Prins GS, Tang WY, Belmonte J, Ho SM. Perinatal exposure to oestradiol and bisphenol A alters the prostate epigenome and increases susceptibility to carcinogenesis. Basic Clin Pharmacol Toxicol. 2008;102:134–8.
13. Yang M, Kim SY, Chang SS, Lee IS, Kawamoto T. Urinary concentrations of bisphenol A in relation to biomarkers of sensitivity and effect and endocrine-related health effects. Environ Mol Mutagen. 2006;47:571–8.
14. Kim M, Bae M, Na H, Yang M. Environmental toxicants-induced epigenetic alterations and their reversers. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2012;30:323–67.
15. Uauy R, Kain J, Corvalan C. How can the Developmental Origins of Health and Disease (DOHaD) hypothesis contribute to improving health in developing countries? Am J Clin Nutr. 2011;94(6 Suppl):1759S–64S.
16. Feinberg AP, Fallin MD. Epigenetics at the crossroads of genes and the environment. JAMA. 2015;314:1129–30.
17. Lee HS, Pyo MY, Yang M. Set, a putative oncogene, as a biomarker for prenatal exposure to bisphenol A. Asian Pac J Cancer Prev. 2012;13:2711–5.
18. Garcia-Arevalo M, Alonso-Magdalena P, Servitja JM, Boronat-Belda T, Merino B, Villar-Pazos S, et al. Maternal exposure to bisphenol-A during pregnancy increases pancreatic beta-cell growth during early life in male mice offspring. Endocrinology. 2016;157:4158–71.
19. Analytical Methods Committee. Using the Grubbs and Cochran tests to identify outliers. Anal Methods. 2015;7:7948–50.
20. Rahman MS, Kwon WS, Yoon SJ, Park YJ, Ryu BY, Pang MG. A novel approach to assessing bisphenol-A hazards using an in vitro model system. BMC Genomics. 2016;17:577.
21. Yang M, Lee HS, Pyo MY. Proteomic biomarkers for prenatal bisphenol A-exposure in mouse immune organs. Environ Mol Mutagen. 2008;49:368–73.
22. Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–61.
23. Pozo K, Castro-Rivera E, Tan C, Plattner F, Schwach G, Siegl V, et al. The role of Cdk5 in neuroendocrine thyroid cancer. Cancer Cell. 2013;24:499–511.
24. Lomnytska MI, Becker S, Gemoll T, Lundgren C, Habermann J, Olsson A, et al. Impact of genomic stability on protein expression in endometrioid endometrial cancer. Br J Cancer. 2012;106:1297–305.
25. Sakwe AM, Koumangoye R, Guillory B, Ochieng J. Annexin A6 contributes to the invasiveness of breast carcinoma cells by influencing the organization and localization of functional focal adhesions. Exp Cell Res. 2011;317:823–37.
26. Vila de Muga S, Timpson P, Cubells L, Evans R, Hayes TE, Rentero C, et al. Annexin A6 inhibits Ras signalling in breast cancer cells. Oncogene. 2009;28:363–77.
27. Lorenz S, Eszlinger M, Paschke R, Aust G, Weick M, Fuhrer D, et al. Calcium signaling of thyrocytes is modulated by TSH through calcium binding protein expression. Biochim Biophys Acta. 2010;1803:352–60.
28. Xiong Q, Zhan S, Zhang N, Ge W, Wang T. Protein profiling of papillary thyroid carcinoma with and without lymph node metastasis: a proteomic study. Int J Clin Exp Pathol. 2016;9:3057–69.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||