AbstractPurposeThe study aimed to search and identify genes that were differentially expressed in breast cancer, and their roles in cancer growth and progression.
Materials and MethodsThe Gene Expression Omnibus (Oncomine) and The Cancer Genome Atlas databases (https://cancergenome.nih.gov/) were screened for genes that were expressed differentially in breast cancer and were closely related to a poor prognosis. Gene expressions were verified by quantitative real-time polymerase chain reaction, and genes were knocked down by a lentivirus-based system. Cell growth and motility were evaluated and in vivo nude mice were used to confirm the in vitro roles of genes. Markers of epithelial-to-mesenchymal transition and the associations of KIF11 with the classical cancer signaling pathways were detected by Western blot.
ResultsA series of genes expressed differentially in patients with breast cancer. The prognosis associated with high KIF11 expression was poor, and the expression of KIF11 increased significantly in high stage and malignant tumor cells. Inhibiting KIF11 expression in lentivirus-suppressed cells revealed that KIF11 inhibition significantly reduced cell viability and colony formation, inhibited migration and invasion, but promoted apoptosis. The sizes and weights of KIF11-inhibited tumors in nude mice were significantly lower than in the negative controls. Western blot showed that E-cadherin in breast cancer was significantly upregulated in KIF-inhibited cells and tumor tissues, whereas N-cadherin and vimentin were significantly downregulated. BT549 and MDA231 cells with KIF11 knockdown exhibited decreased ERK, AMPK, AKT, and CREB phosphorylation.
IntroductionBreast cancer is the most common and the fifth most common cause of cancer-related death worldwide; however, it is the leading cause of cancer-related death among females in less-developed countries [1,2]. In 2017 alone, the American Cancer Society reported 252,710 new cases, of which 40,610 cases died because of the malignancy. Between 2005 and 2014, the prevalence of breast cancer in the Asia-Pacific region showed an annual growth rate of approximately 1.7% [3]. In China, it is now estimated that there are 278,800 newly diagnosed cases and 64,600 deaths per year [1]. Studies focusing on the etiology of breast cancer are of great importance if we are to improve early diagnosis and provide targeted therapy.
Although the pathology of breast cancer is not fully understood, many genes are thought to be involved. For instance, ADHFE1 functions as a breast cancer oncogene that decreases patient survival, and when it is overexpressed with MYC, it can enhance tumor growth in vitro [4]. SOX2, which belongs to a family of high-mobility group transcription factors, is frequently overexpressed in breast cancer tissues and plays an important role in cell proliferation and mammary tumorigenesis [5]. Skp2, which is highly expressed in breast cancer specimens, is significantly associated with poor clinical outcome [6]. Large-scale genomic studies using wholegenome small hairpin RNA have also revealed that BRD4 may be a potential anticancer target [7]. Finally, overexpressed and amplified IKBKE has been shown to function as an oncogene, with suppressed expression leading to cell death in breast cancer [8].
Despite the progress in the research on the causative genes related to breast cancer progression, there remain a large number of genes that have not been fully characterized in breast cancer. Therefore, we aimed to search and identify genes that were differentially expressed in breast cancer, before verifying their roles in cancer growth and progression.
Materials and Methods1. Cell cultureWe cultured BT474, BT549, MCF-1, MDA231, and SK-BR-3 in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS; HyClone, Logan, UT) and incubated at 37°C in a 5% CO2 atmosphere. All cell lines were purchased from the Shanghai Cell Bank of Chinese Academy of Medical Sciences (Shanghai, China).
2. Quantitative real-time polymerase chain reactionTotal RNA was isolated from breast cells using TRIzol (Thermo Fisher Scientific, Waltham, MA) following a standard protocol. The reverse transcription products of total RNA from each sample were obtained by using the Prime-Script RT reagent kit (Takara, Tokyo, Japan). The SYBR Real-time PCR Master Mix kit (TOYOBO, Osaka, Japan) was used for mRNA amplification, according to the manufacturer's protocol, and the amplification reactions were run on an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA). Data generated by quantitative real-time polymerase chain reaction were analyzed by the 2-ΔΔCt method. Each reaction was set up with three replicates per group.
3. Lentivirus-mediated RNA interferenceTwo small hairpin RNA of KIF11 (siKIF11-1, sense 5′-CCACGTACCCTTCATCAA-3′; siKIF11-2, sense 5′-GGAGGAGCTGAATAGGGTT-3′) and a negative control (NC; sense 5′-UUCUCCGAACGUGUCACGUTT-3′) were cloned to the HpaI and XhoI restriction sites of the LV-003 vector (Forevergen, Guangzhou, China), respectively, to generate KIF11 interference plasmids. The resulting LV-003 siKIF11-1 and LV-003 siKIF11-2 vectors were co-transfected with a packaging vector into HEK-293T cells. At 48 and 72 hours after transfection, lentivirus was harvested from the cell supernatant by ultracentrifugation at 25,000 revolutions per minute for 2 hours. The lentivirus was suspended in phosphate-buffered saline and subjected to cell infection. Stable KIF11-knockdown cell lines, siKIF11-1 and siKIF11-2, were cultured in a medium supplemented with 2 μg/mL puromycin for 48 hours.
4. Western blotCells were analyzed with the RIPA buffer (Pierce, Rockford, IL) on ice and were harvested. Cell lysate protein was quantified by the BCA Protein Assay Kit (Pierce) and separated on 12% polyacrylamide gels, which were then transferred to PDVF membranes (Millipore, Billerica, MA) and blocked with 5% non-fat milk Tris-buffered saline. Thereafter, the membranes were incubated overnight at 4°C in Eg5 primary antibodies (ab37814, 1:250 dilution, Abcam, Cambridge, MA) followed by room temperature for 1 hour in horseradish peroxidase-conjugated secondary antibodies of Goat Anti-Rabbit IgG (ab6721, 1:2,000 dilution, Abcam). Blots on the membranes were visualized by chemiluminescence (Pierce), with glyceraldehyde 3-phosphate dehydrogenase used as an internal reference control.
5. MTS assayTwelve hours before MTS assay with 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, the cell suspension was seeded onto a 96-well plate at a density of 1×104. DMEM (100 μL) supplemented with 20 μL of Cell Titer 96 Aqueous One Solution Reagent (Promega, Madison, WI) was added to each well to determine cell viability. After 1, 2, and 3 days of culture, cell viability was quantified by reading the absorbance at 490 nm on a 96-well plate reader (Bio-Rad Laboratories, Hercules, CA).
6. CytometryTo evaluate the apoptosis rate in the breast cells, 1×106 cells were suspended in phosphate-buffered saline and stained with 5 μL of annexin V and 1 μL of propidium iodide, according to the manufacturer's instructions. After 15 minutes of incubation, the stained cells were subjected to BD LSR II flow cytometry (BD Biosciences, San Jose, CA) with the FlowJo software, and we manually analyzed apoptosis.
7. Colony formation assayCells were seeded in 6-well culture dishes, at densities of 100, 200, and 400 cells/well. After 2 weeks, cell colonies that formed on the plates were fixed with 4% paraformaldehyde and visualized by staining with 0.1% crystal violet. Cell colonies that formed in each group from three independent experiments were counted and expressed as clone formation ratios.
8. Transwell assayApproximately 1×105 cells were seeded in the upper compartment of a transwell (BD Biosciences) and cultured in serum-free DMEM medium for the assessment of cell migration and invasion. The lower chambers were added with DMEM medium containing 10% FCS. In the migration assay, cells in the upper compartment migrated through a micrometer pore filter to the lower chamber, whereas cells migrated through a Matrigel-coated filter in the invasion assay. The cell numbers in the lower compartment were counted per group under an inverted microscope with 0.1% crystal violet staining. Three high quantitative microscopic views were selected for calculation. All experiments were performed in triplicate.
9. Xenograft mouse tumorsA total of ten 6-week-old BALB/c nu/nu mice were randomly assigned to either a siKIF11 group or a NC group. In the siKIF11 group, cells transduced with siKIF lentivirus were implanted subcutaneously into nude mice and monitored weekly for 5 weeks, and the tumor volume (volume [mm3]=length×width2×0.5) and tumor weight were recorded every week. The NC mice were implanted with lentivirus cells. The animal experiments were approved by the Institutional Research Medical Ethics Committee of the First Affiliated Hospital of Xiamen University.
10. Bioinformatics and statistical analysisTo explore the mRNA profiling of breast cancer, the expression pattern of KIF11, and the relative expressions of KIF11 in different cancers, data retrieved from Oncomine were analyzed using their web portal (http://www.oncomine.org). Analyses of the difference in KIF11 expression between breast tumors and paired/unpaired adjacent normal tissues, as well as the overall and recurrence-free survival times of the KIF11-high-expression patients, were analyzed on the basis of the data downloaded from The Cancer Genome Atlas (https://cancergenome.nih.gov/), using the Oncomine online analysis tools [9]. Data are expressed as means±standard deviation. The Student's t test was used to analyze the difference between two groups. One-way analysis of variance followed by the least significant difference test was used to analyze the differences among more than two groups. For survival analyses, Kaplan-Meier curves were generated and the differences between groups were analyzed by log-rank tests. A p-value of < 0.05 was considered statistically significant. The database GEPIA (http://gepia.cancer-pku.cn/) was used to generate the pathological stage plot of KIF11.
Results1. Screening differential genes involved in breast cancer progressionTo screen candidate genes that may be associated with breast cancer pathology, data sets were retried from Oncomine. As shown in Fig. 1A, COL11A1, GJB2, MMP11, MMP13, PPAPDC1A, TPX2, SPAGS, HAGHL, KIF11, and CST2 were the 10 most upregulated genes, followed by LACE3, POC1A, TPX2, SCT, EPYC, POP1, RACGAP1, BMP8A, C6orf129, DDX39, NVL, and HAGHL. We selected LACE3, KIF11, POP1, NVL, PARN, CCDC167 (C6orf129), PPAPDC1A, and DDX39 for prognostic evaluation because their functions remain unknown in breast cancer. The cut-off for high/low expression is 50%. As shown in Fig. 1B, patients with breast cancer who had high expressions of POP1, PPAPDC1A, and KIF11 had shorter overall survival times than those with lower expressions. By contrast, patients with high expressions of LACE3, NVL, PARN, CCDC167 (C6orf129), and DDX39 showed better overall survival compared with those who had lower expressions. Among these genes, KIF11, POP1, and PPAPDC1A were most significantly different and were included in the further analyses (Fig. 1B).
2. KIF11 overexpression was associated with poor survival times in patients with breast cancerBy analyzing the expression levels of KIF11, POP1, and PPAPDC1A in 19 tumors and adjacent normal controls using the Fire browse database, they were each upregulated in almost all tumor tissues, including breast cancer (Fig. 2). To verify the expression levels of these three genes in breast cancer, differences in their expression levels were analyzed in The Cancer Genome Atlas database using the information of tumor tissue and paired/unpaired cancer adjacent controls. The results showed that compared with either the paired paracancer (Fig. 3A) or the unpaired paracancer control (Fig. 3B), the expression levels of KIF11, POP1, and PPAPDC1A were significantly upregulated in tumor tissues (Fig. 3). Analysis of the relationship between KIF11, POP1, and PPAPDC1A expression and The survival time of patients with breast cancer in The Cancer Genome Atlas database revealed that the overall survival of those with high KIF11 expression was significantly lower than that of patients with low expression (Fig. 4A). In addition, analysis of recurrence-free survival showed that patients with high KIF11 and POP1 expressions had high recurrence rates (Fig. 4B). Together, these findings indicated that overexpressed KIF11 occurred frequently in breast cancer and was associated with a poor prognosis.
High KIF11 expression being associated with a poor prognosis in patients led us to study the potential function of KIF11 in breast cancer progression. First, we identified KIF11 expression in both tumorous and normal cell lines. Analysis of KIF11 expression in 25 tumor tissues of patients with different stages of breast cancer revealed that the expression level of KIF11 in patients with stage II-B/III disease (n=11) was significantly higher than that in patients with stage I-A/II-A disease (n=14) (Fig. 5A). Then the pathological stage plot showed that the KIF11 expression levels differ between different stages of cancer (p < 0.05) (S1 Fig.). In addition, analysis of the background expression of KIF in five breast cancer cell lines (i.e., BT474, BT549, MCF-1, MDA231, and SK-BR-3) revealed that KIF11 was highly expressed in two breast cancer cells with high metastatic potential (i.e., BT549 and MDA231) (Fig. 5B).
3. Knockdown of KIF11 inhibits breast cancer cell growth and metastasis in vitroTo study the role of KIF11 in breast cancer cell biology, KIF11 knockdown was performed by a lentivirus-based system in both BT549 and MDA231 cells. Two small hairpin RNA were designed for KIF11 downregulation in both cell lines to generate KIF11 knockdown stable cell lines of MDA-231 (i.e., MDA231 siKIF-1 and MDA231 siKIF-2) and BT549 (i.e., BT549 siKIF-1 and BT549 siKIF-2). The results of qRTPCR showed that KIF11 was significantly downregulated in both MDA231 siKIF-1 and MDA231 siKIF-2 compared with the NC MDA231 LV003 cells. KIF11 downregulation was also observed in the BT549 siKIF-1 and the BT549 siKIF-1 cells compared with the BT549 LV003 cells (Fig. 5C). Western blot confirmed the downregulation of KIF11 in both the BT549 and MDA231 cells (Fig. 5D). We selected MDA231 siKIF-2 and BT549 siKIF-2 cells for the subsequent functional analysis.
To investigate the effects of KIF11 downregulation on cell growth, we assessed viability, apoptosis, and colony formation by MTS, cytometry, and colony formation assays. MTS assay showed that KIF11 knockdown significantly impaired cell viability in both the BT549 and MDA231 siKIF-2 cells compared with those cells receiving no treatment (Mock) and corresponding NCs after 3 days of culture (Fig. 5E). The results of MTS also showed that KIF11 knockdown significantly impaired cell viability in both the BT474 and SK-BR-3 siKIF-2 cells compared with those cells receiving no treatment (Mock) and corresponding NCs after 3 days of culture (S2 Fig.). Cytometry results showed much higher apoptotic cell proportion rate in the BT549 and MDA231 siKIF-2 cells than that in the NC cells (Fig. 6A). Furthermore, colony formation assay displayed that regardless of the number of initial cell cultures of 100, 200, or 400, the number of colony formations in the BT549 and MDA231 siKIF-2 cells were less than those in the NC cells (Fig. 6B).
The effects of KIF11 downregulation on breast cell migration and invasion were evaluated both in vitro and in vivo. Cells cultured in the upper transwell compartment migrated through 8-μm holes during the migration assay or through Matrigel-coated holes during the invasion assay. As shown in Fig. 6, the KIF11 knockdown cell (siKIF11) showed a significant decrease in the number of migrated BT549 cells in both the migration and invasion assays compared with NC cells. Similar trends were observed in the MDA231 siKIF11 knockdown cells, with fewer siKIF cells than NC cells in the lower chamber (Fig. 6C).
4. KIF11 knockdown inhibited breast cancer cell progression in nude miceTo confirm the functional effect of KIF11 observed in vitro, BT549 siKIF11 cells were implanted subcutaneously into nude mice and determined weekly to generate a tumor growth curve. The curve for tumor volume showed that mice bearing KIF11-downregulated tumors had significantly smaller tumor volumes compared with the NC mice at each observation. Moreover, the tumors did not grow over 5 weeks in the siKF11 mice, but they grew continuously in the NC mice (Fig. 7A and C). After 5 weeks, tumors in the mice implanted with BT549 siKIF11 cells were almost undetectable, whereas those in the mice with NC cells were clearly visible (Fig. 7A). The siKIF11 mice also had significantly lower tumor weight compared with the NC mice (Fig. 7B). Western blotting confirmed that KIF11 was significantly downregulated in the siKF11 mice (Fig. 7D). Hematoxylin and eosin staining showed that the cells were arranged neatly with good morphological consistency in mammary gland sections of the siKIF11 mice compared with the NC mice (Fig. 7E). Tumor cells in the NC group proliferated faster to necrosis and were denser compared with the siKIF11 group.
5. The epithelial-to-mesenchymal transition pathway is involved in the regulatory role of KIF11To study whether the epithelial-to-mesenchymal transition (EMT) pathway is involved in the observed effect of KIF11 downregulation, markers of EMT (i.e., E-cadherin, N-cadherin, and vimentin) were measured by Western blot in both the siKIF11 or normal BT549 and MDA231 cells. Compared with the NC BT549 cells, the protein expression of E-cadherin was significantly enhanced and the expressions of N-cadherin and vimentin were downregulated in the siKIF11 cells. Consistent with the results observed in the BT549 cells, siKIF11 MDA231 cells showed higher expressions of E-cadherin and lower expressions of N-cadherin and vimentin compared with the NC mice (Fig. 8A).
6. Modulated signaling in BT549 and MDA231 cells by KIF11 knockdownFor more insights into the cell functions regulation by KIF11 during breast cancer progression, multiple key signaling pathways components (ERK, AMPK, AKT, and CREB) associated with cell proliferation, migration, invasion, cell cycle and apoptosis were further measured by western blot in siKIF11 or normal BT549 and MDA231 cells. Here we showed that the phosphorylated ERK, AMPK, AKT, and CREB level was significantly inhibited in BT549 and MDA-231 cells with KIF11 knockdown (Fig. 8B). These alterations of key signaling components suggested that regulation of breast cancer cell proliferation, migration, invasion, and cell cycle progression by KIF11 might be at least partially mediated by modulating the phosphorylation of ERK, AMPK, AKT, and CREB, suggesting the activation of the corresponding pathways such as mitogen-activated protein kinase (MAPK)/ERK and phosphoinositide 3-kinase (PI3K)/AKT pathway.
DiscussionIn this study, we identified that high KIF11 expression was associated with poor overall and recurrence-free survival. Of note, KIF11 was upregulated in patients with higher grade disease and with malignant breast cancer cell lines. When we downregulated this expression, we observed inhibitory effects on cell proliferation, colony formation, invasion, and migration, as well as an enhancement of apoptosis. In vivo study of KIF11 knockdown cells implanted in mice confirmed that KIF11 downregulation ameliorated tumor progression and contributed to a change in the cell phenotype from epithelial to mesenchymal.
KIF11, or kinesin family member 11, belongs to the kinesin-5 family and is thought to have important functions in tetrameric microtubule cross-linkage, cell mitosis, and the cell cycle and differentiation [10,11]. Studies have suggested that a genetic variant of KIF11 is closely related to familial exudative vitreoretinopathy [12,13], congenital lymphedema, and chorioretinopathy [14] as well as to an increased risk of type 2 diabetes [15]. In cancer, although the role of KIF11 has not been fully elucidated, studies have shown that it is overexpressed in many cancers. For instance, Imai et al. [16] sugges-ted that KIF11 was frequently expressed in gastric cancer exhibiting the intestinal phenotype. In bladder, renal cell, and astrocytic cancers, KIF11 overexpression has been associated with cancer stage and recurrence [17-19]. In laryngeal squamous cell carcinoma, high KIF11 expression is correlated to lymph node metastasis, TNM stage, and an unfavorable prognosis [20]. In oral cancer, significant KIF11 upregulation is correlated with a shorter survival time and may be an independent prognostic factor [21]. Here we showed that KIF11 was upregulated in breast cancer when compared with either paired or unpaired adjacent cancer tissues. Moreover, we confirmed that the expression of KIF11 was greater in patients with higher clinical stages. The online data sets revealed that KIF11 overexpression in patients with breast cancer was associated with poor outcomes. Together, these findings indicate that a close relationship exists between KIF11 and breast cancer.
In tandem with the negative effects of KIF11 overexpression, we validated the beneficial role of KIF11 downregulation on tumor growth and metastasis via a lentivirus-based system. This supported the idea that KIF11 could represent a viable therapeutic target. Previously, several studies have suggested that KIF11 is important to cell proliferation and motility in various cancers, including breast cancer. The overexpression of KIF11 has been shown to result in enhanced anchorage-independent cell growth and tumor formation in a highly aggressive pancreatic tumor model [22]. In turn, the inhibition of KIF11 was shown to impair the cell growth and motility of glioblastoma cells and to prolong the survival time of orthotopic tumors in mice [23]. In pancreatic cancer, the inhibition of KIF11 by dimethylenastron has also been reported to affect cell migration and invasion [24].
Concerning breast cancer, several studies have reported the therapeutic effect of KIF11 inhibition. KIF11 was reported to be involved in fulvestrant-mediated G0/G1 phase arrest of the cell cycle in estrogen-positive MCF-7 cells. The blockade of KIF11 by this estrogen receptor inhibitor resulted in decreased cell growth [25]. Administration of 3,4-dihydropyrimidin-2(1H)-one (or thione) derivatives, which are KIF11 inhibitors that modify the motor ability of KIF11, were shown to affect the phenotype of cancer stem cells critical to breast cancer initiation and development. This impaired proliferation and induced apoptosis in MCF-7 and MDA231 cells, and it induced angiogenesis in vitro and in vivo [26]. Another KIF11 inhibitor, K85, was shown to have anticancer activity in breast cancer, though it also contributed to chemoresistance [27]. Adding to this existing research, we showed that there was a clinical association between the expression of KIF11 and breast cancer. Moreover, we validated that KIF11 promoted tumor growth and metastasis in BT549 and MDA-231 cells in a lentivirus-based system. We are aware of no prior data on the role of KIF11 in BT549 cells.
Of note, our research indicated that the EMT process was involved in the observed effect of KIF11 on breast cancer progression both in vivo and in vitro. The EMT process involves cells gradually losing their epithelial phenotype and acquiring a mesenchymal phenotype. Thus, EMT can lead to disordered cell-to-cell adhesion and junctions, allowing cells to obtain motility and potentially cross the extracellular matrix to achieve distant metastasis [28]. In this study, we observed downregulated expression of mesenchymal markers (N-cadherin and vimentin) and elevated expression of an epithelial marker (E-cadherin), indicating a reversed EMT process. Recently, the relationship between EMT and KIF11 has been revealed in glioblastoma cells, with KIF11 knockdown using K858 rescuing the mesenchymal phenotype of cells [29]. We propose that KIF11 knockdown reduces the motility of breast cancer cells by promoting the epithelial rather than the mesenchymal phenotype, thereby inhibiting cancer metastasis and recurrence. In the study, multiple key signaling pathways components associated with cell proliferation, migration, invasion, cell cycle and apoptosis in BT549 and MDA-231 cells transfected with KIF11 siRNAs were further measured by western blotting. The significantly altered ERK, AMPK, AKT, and CREB phosphorylation further validated the critical roles played by KIF11 in breast cancer cells. It has been proved that several signaling pathways have been shown to be implicated in tumorigenesis, which include the JAK/STAT pathway, the NOTCH signaling pathway, the Wnt pathway, the MAPK/ERK pathway, the PI3K/AKT pathway, the nuclear factor кB pathway, and the transforming growth factor β pathway [30-32]. Inappropriate cell survival and uncontrolled proliferation are among the characteristics of cancer, and these processes are commonly regulated by the MAPK/ERK pathway [33,34]. Protein kinase B (AKT) is another key regulator of tumor cell proliferation, cell cycle progression, migration and invasion, which is also activated by its phosphorylation [35-37]. The results in our current study suggested that KIF11 might affect MAPK/ERK and PI3K/AKT signaling pathway by interacting with specific kinase or phosphatase. Further study should be conducted to elucidate the associations of KIF11 with these complex signaling networks.
In conclusion, we have reported on the relationships between the expressions of several genes and breast cancer survival based on the Oncomine online tool. Analysis revealed that siKIF11 had a role in suppressing tumor growth and metastasis in both BT549 and MDA231 cells, as well as in nude mice with BT549 allograft tumors. We posit that this occurred through regulation of the EMT process and affecting signaling pathways that are known to promote the development and progression of cancer. The present findings support the idea that KIF11 is an oncogene and may be targeted in the treatment of breast cancer.
Electronic Supplementary MaterialSupplementary materials are available at Cancer Research and Treatment website (http://www.e-crt.org).
AcknowledgmentsThis work was partly supported by the National Natural Science Foundation of China (81802600, 81872459, 81803050), the Natural Science Foundation of Fujian Province (2016J01635), the Natural Science Foundation of Guangdong Province (2018A030313666), and the Science and Technology Planning Projects of Xiamen Science & Technology Bureau (3502Z20174070).
Fig. 1.Analysis of differentially expressed genes in patients with breast cancer. (A) Heat map of relative mRNA expression (normalized to tumor adjacent normal tissues) in patients with breast cancer, analyzed by the Oncomine database online tool. (B) Survival analyses of patients with breast cancer and different expression levels of LAGE3, KIF11, POP1, PARN, CCDC167, DDX39, NVL, and PPAPDC1A using the OncoLnc online survival analysis tool (http://www.oncolnc.org/). 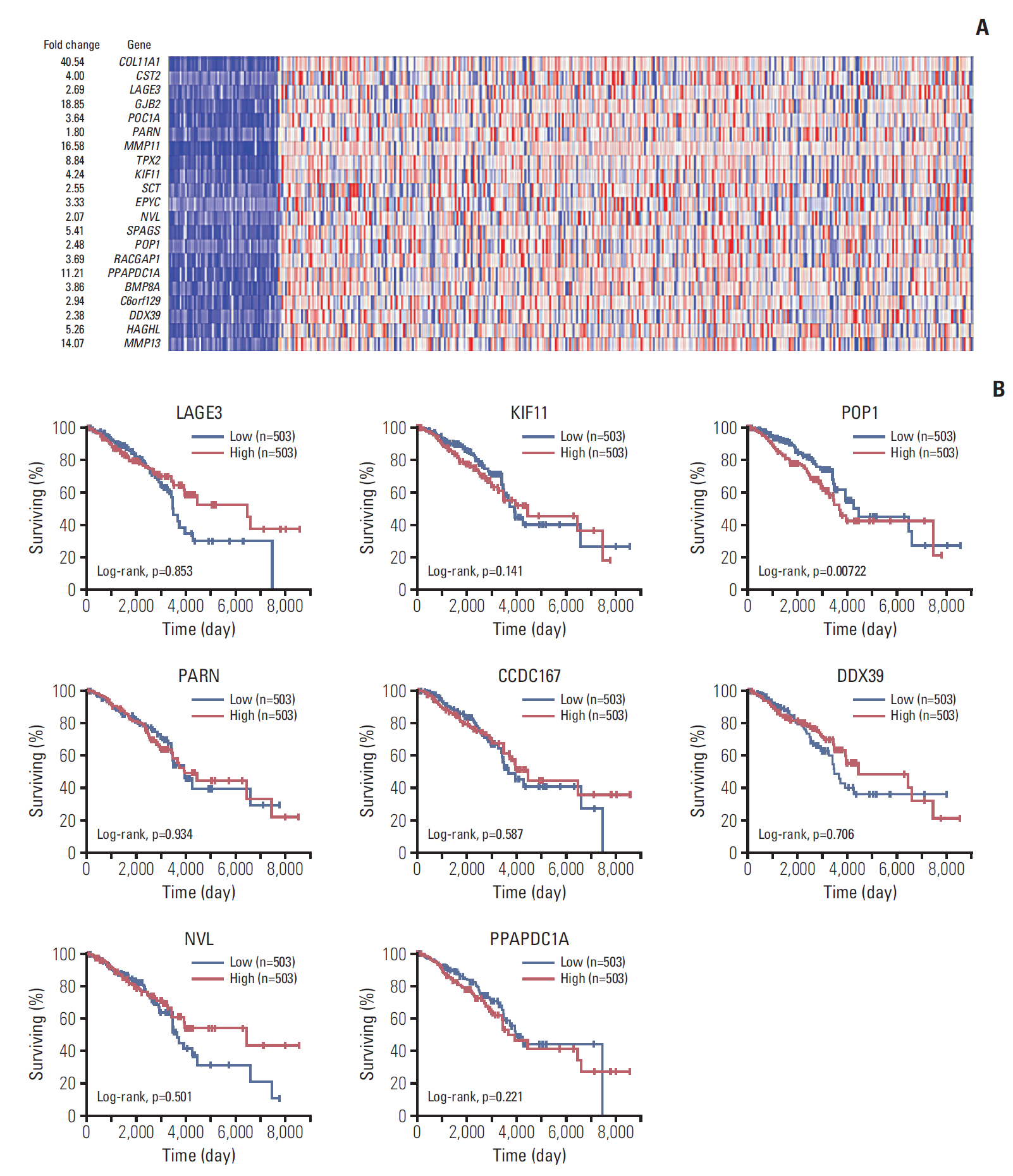
Fig. 3.Expression analyses of KIF11, POP1, and PPAPDC1A in breast cancer based on The Cancer Genome Atlas. (A) The relative expression of KIF11, POP1, and PPAPDC1A in breast cancer versus non-paired tumor adjacent normal tissues. ****p < 0.0001. (B) The relative expressions of KIF11, POP1, and PPAPDC1A in breast cancer versus paired tumor adjacent normal tissues. 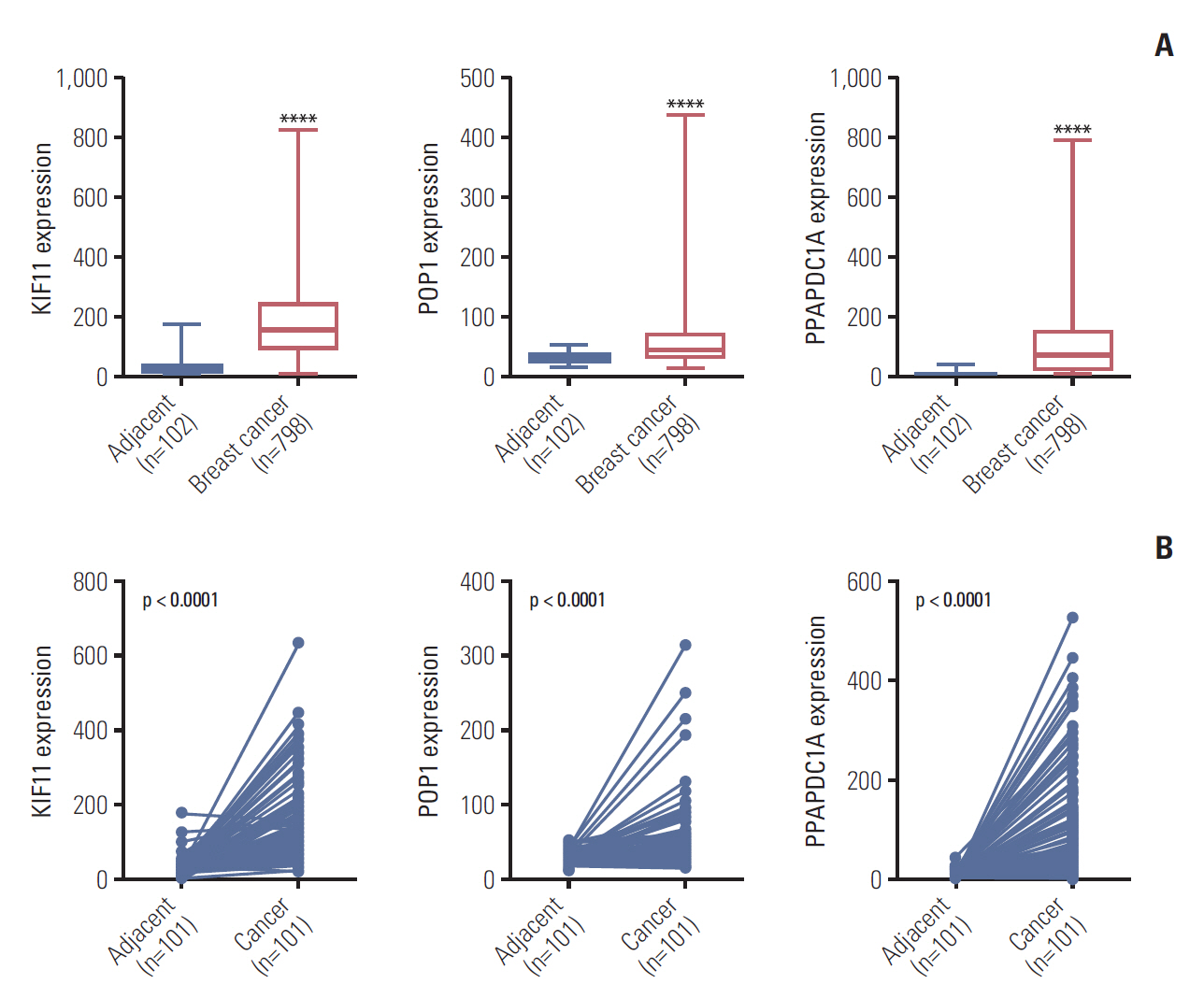
Fig. 4.Overall survival (OS) (A) and recurrence-free survival (RFS) (B) in patients with breast cancer by low and high KIF11, POP1, and PPAPDC1A expressions based on The Cancer Genome Atlas. HR, hazard ratio; CI, confidence interval. 
Fig. 5.Analysis of KIF11 in breast cancer tissue and cells. (A) KIF11 expression in patients with breast cancer of grades I-1/II-A and II-B/III. (B) The relative background expressions of KIF11 in six breast cancer cell lines. (C) Both the BT549 and the MDA231 cell line were transduced with lentivirus (Lv003), siKIF11-1, or siKIF11-2 as negative controls, and the relative mRNA levels of KIF11 were detected by quantitative real-time polymerase chain reaction. (D) Protein levels of KIF11 in BT549 and MDA231 cells were detected by Western blotting. (E) Cell viability was evaluated by MTS in both BT549 and MDA231 cells. NC, cells transduced with empty Lv003 lentivirus; MOCK, cells without any treatment; siKIF11-2, cells transduced with siKIF11-2 lentivirus. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. 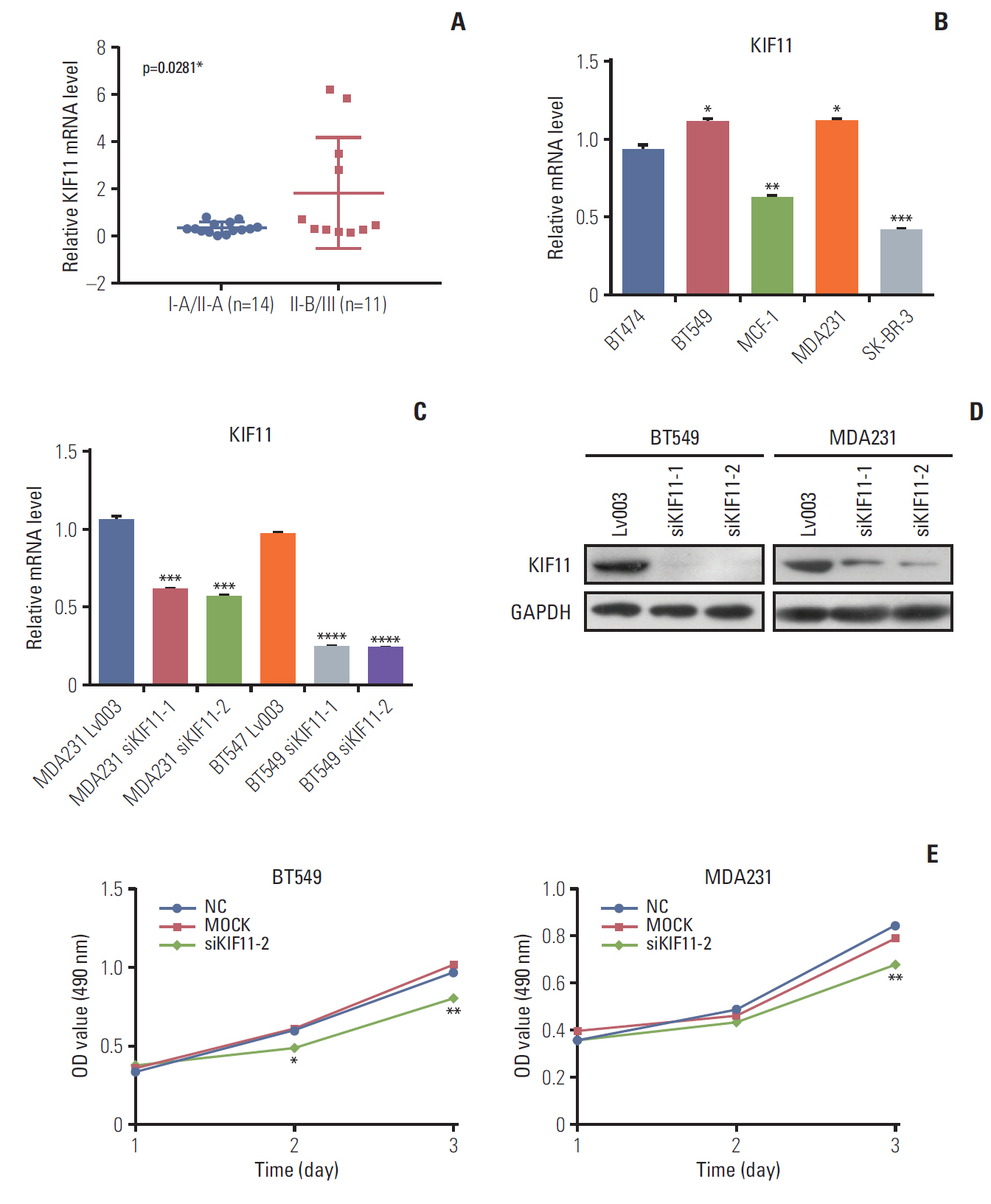
Fig. 6.Downregulation of KIF11 influenced cell apoptosis, cell proliferation, cell migration, and invasion. (A) Cell apoptosis was determined by cytometry in KIF11-suppressed BT549 and MDA231 cells. (B) Cell proliferation of the KIF11-suppressed BT549 and MDA231 cells was assessed by clone forming assays. (C) Downregulation of KIF11 inhibited the cell migration and invasion capabilities of the BT549 and MDA231 cells. NC, normal control. *p < 0.05, **p < 0.01. 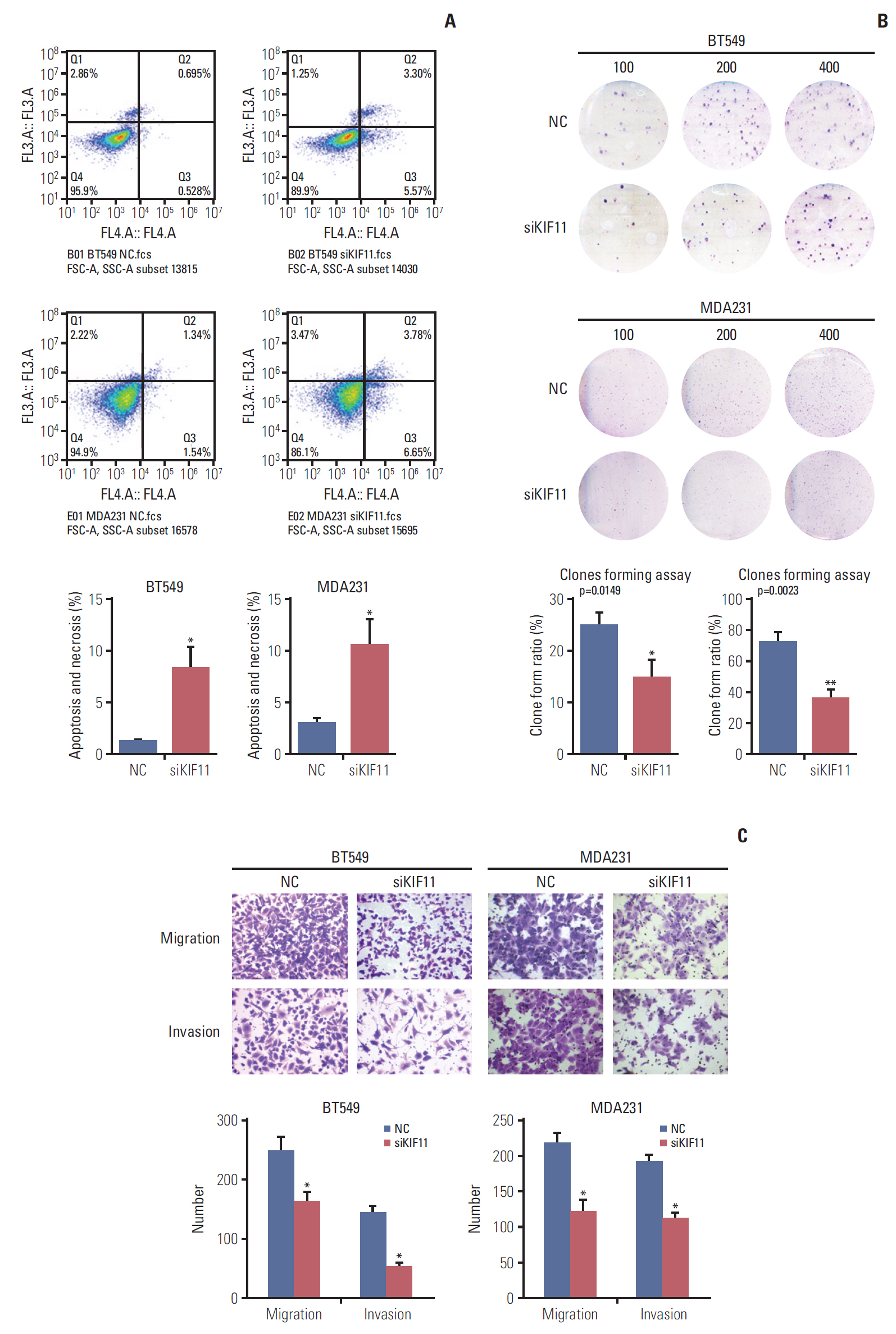
Fig. 7.Downregulation of KIF11 inhibited tumor growth in xenograft BALB/c nu/nu mice. (A) Appearance at 5-week post-implantation of BALB/c nu/nu mice xenografted with BT549 cells that had either normal (NC) or low KIF11 (siKIF) expression. (B) The tumor volume curve in each group between days 10 and 35 post-implantation. (C) The tumor weight in each group determined at 5 weeks after implantation. (D) Western blotting confirmed the downregulation of KIF11 in siKIF11 mice. (E) Representative image of hematoxylin and eosin showing the beneficial effect of KIF11 downregulation in breast tumors at 5-week post-implantation (magnification, 100×). GAPDH, glyceraldehyde 3-phosphate dehydrogenase. *p < 0.05, **p < 0.01, ****p < 0.0001. 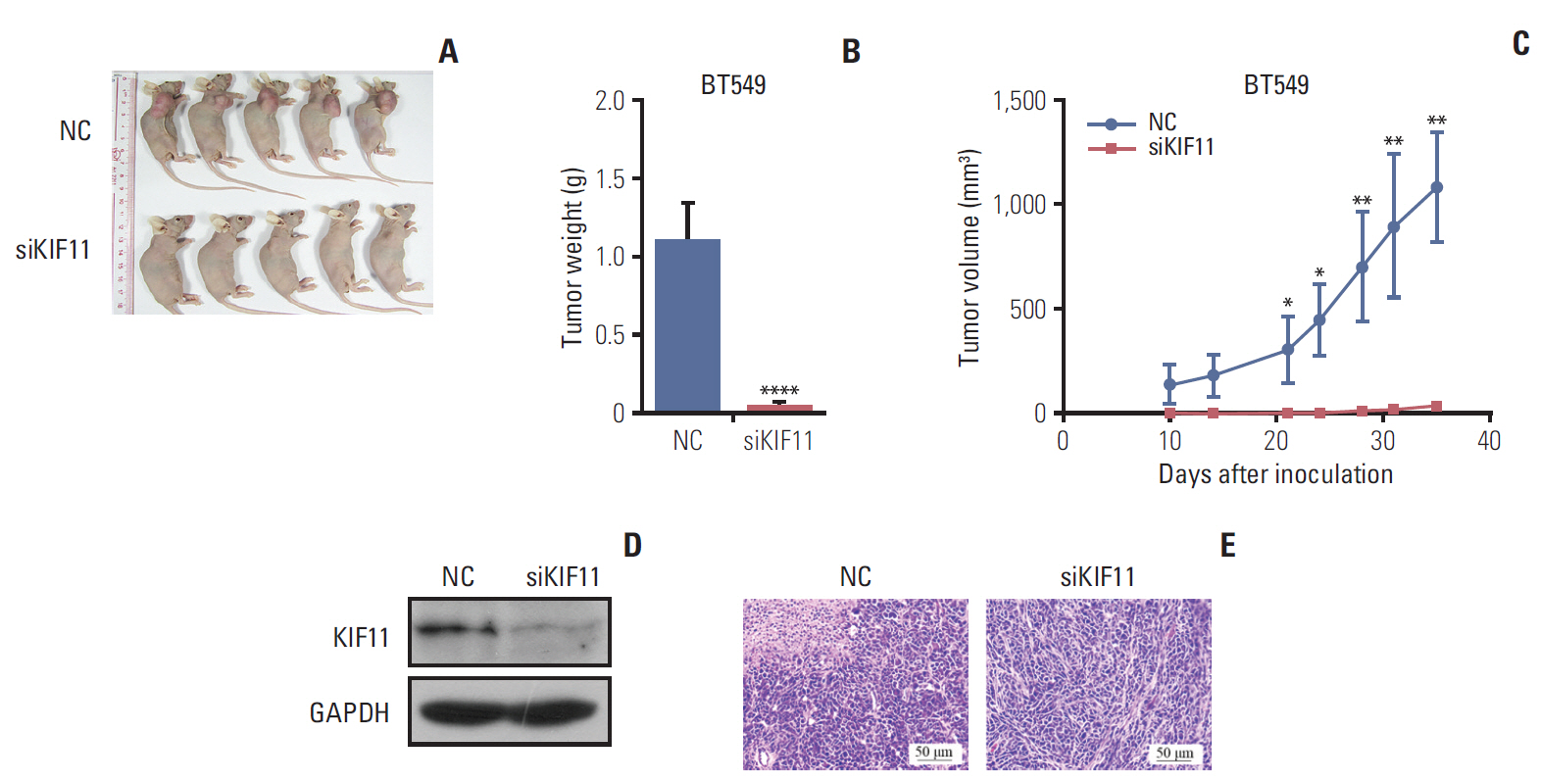
Fig. 8.Representative Western blot showing the expressions of markers of epithelial-to-mesenchymal transition (EMT) and the associations of KIF11 with the classical cancer signaling pathways. (A) Markers of EMT (E-cadherin, N-cadherin, and vimentin) were measured by Western blot in both the siKIF11 or normal BT549 and MDA231 cells. (B) Multiple key signaling pathways components (ERK, AMPK, AKT, and CREB) were measured by western blot in both the siKIF11 or normal BT549 and MDA231 cells. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal standard. 
References1. Zuo TT, Zheng RS, Zeng HM, Zhang SW, Chen WQ. Female breast cancer incidence and mortality in China, 2013. Thorac Cancer. 2017;8:214–8.
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32.
3. DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67:439–48.
4. Mishra P, Tang W, Putluri V, Dorsey TH, Jin F, Wang F, et al. ADHFE1 is a breast cancer oncogene and induces metabolic reprogramming. J Clin Invest. 2018;128:323–40.
5. Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, et al. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem. 2008;283:17969–78.
6. Traub F, Mengel M, Luck HJ, Kreipe HH, von Wasielewski R. Prognostic impact of Skp2 and p27 in human breast cancer. Breast Cancer Res Treat. 2006;99:185–91.
7. Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, et al. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164:293–309.
8. Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–79.
9. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80.
10. Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T. Directional switching of the kinesin Cin8 through motor coupling. Science. 2011;332:94–9.
11. Ma HT, Erdal S, Huang S, Poon RY. Synergism between inhibitors of Aurora A and KIF11 overcomes KIF15-dependent drug resistance. Mol Oncol. 2014;8:1404–18.
12. Hu H, Xiao X, Li S, Jia X, Guo X, Zhang Q. KIF11 mutations are a common cause of autosomal dominant familial exudative vitreoretinopathy. Br J Ophthalmol. 2016;100:278–83.
13. Rao FQ, Cai XB, Cheng FF, Cheng W, Fang XL, Li N, et al. Mutations in LRP5,FZD4, TSPAN12, NDP, ZNF408, or KIF11 genes account for 38.7% of Chinese patients with familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2017;58:2623–9.
14. Mears K, Bakall B, Harney LA, Penticoff JA, Stone EM. Autosomal dominant microcephaly associated with congenital lymphedema and chorioretinopathy due to a novel mutation in KIF11. JAMA Ophthalmol. 2015;133:720–1.
15. Hu C, Zhang R, Wang C, Wang J, Ma X, Lu J, et al. PPARG, KCNJ11, CDKAL1, CDKN2A-CDKN2B, IDE-KIF11-HHEX, IGF2BP2 and SLC30A8 are associated with type 2 diabetes in a Chinese population. PLoS One. 2009;4:e7643
16. Imai T, Oue N, Nishioka M, Mukai S, Oshima T, Sakamoto N, et al. Overexpression of KIF11 in gastric cancer with intestinal mucin phenotype. Pathobiology. 2017;84:16–24.
17. Wissing MD, De Morree ES, Dezentje VO, Buijs JT, De Krijger RR, Smit VT, et al. Nuclear Eg5 (kinesin spindle protein) expression predicts docetaxel response and prostate cancer aggressiveness. Oncotarget. 2014;5:7357–67.
18. Sun D, Lu J, Ding K, Bi D, Niu Z, Cao Q, et al. The expression of Eg5 predicts a poor outcome for patients with renal cell carcinoma. Med Oncol. 2013;30:476.
19. Liu L, Liu X, Mare M, Dumont AS, Zhang H, Yan D, et al. Overexpression of Eg5 correlates with high grade astrocytic neoplasm. J Neurooncol. 2016;126:77–80.
20. Lu M, Zhu H, Wang X, Zhang D, Xiong L, Xu L, et al. The prognostic role of Eg5 expression in laryngeal squamous cell carcinoma. Pathology. 2016;48:214–8.
21. Daigo K, Takano A, Thang PM, Yoshitake Y, Shinohara M, Tohnai I, et al. Characterization of KIF11 as a novel prognostic biomarker and therapeutic target for oral cancer. Int J Oncol. 2018;52:155–65.
22. Liu M, Wang X, Yang Y, Li D, Ren H, Zhu Q, et al. Ectopic expression of the microtubule-dependent motor protein Eg5 promotes pancreatic tumourigenesis. J Pathol. 2010;221:221–8.
23. Venere M, Horbinski C, Crish JF, Jin X, Vasanji A, Major J, et al. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci Transl Med. 2015;7:304ra143.
24. Sun XD, Shi XJ, Sun XO, Luo YG, Wu XJ, Yao CF, et al. Dimethylenastron suppresses human pancreatic cancer cell migration and invasion in vitro via allosteric inhibition of mitotic kinesin Eg5. Acta Pharmacol Sin. 2011;32:1543–8.
25. Planas-Silva MD, Filatova IS. Estrogen-dependent regulation of Eg5 in breast cancer cells. Anticancer Drugs. 2007;18:773–9.
26. Guido BC, Ramos LM, Nolasco DO, Nobrega CC, Andrade BY, Pic-Taylor A, et al. Impact of kinesin Eg5 inhibition by 3,4-dihydropyrimidin-2(1H)-one derivatives on various breast cancer cell features. BMC Cancer. 2015;15:283.
27. De Iuliis F, Taglieri L, Salerno G, Giuffrida A, Milana B, Giantulli S, et al. The kinesin Eg5 inhibitor K858 induces apoptosis but also survivin-related chemoresistance in breast cancer cells. Invest New Drugs. 2016;34:399–406.
28. Creighton CJ, Chang JC, Rosen JM. Epithelial-mesenchymal transition (EMT) in tumor-initiating cells and its clinical implications in breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:253–60.
29. Taglieri L, Rubinacci G, Giuffrida A, Carradori S, Scarpa S. The kinesin Eg5 inhibitor K858 induces apoptosis and reverses the malignant invasive phenotype in human glioblastoma cells. Invest New Drugs. 2018;36:28–35.
30. Wang TL, Rago C, Silliman N, Ptak J, Markowitz S, Willson JK, et al. Prevalence of somatic alterations in the colorectal cancer cell genome. Proc Natl Acad Sci U S A. 2002;99:3076–80.
31. Weng AP, Ferrando AA, Lee W, Morris JP 4th, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71.
32. Armstrong L, Hughes O, Yung S, Hyslop L, Stewart R, Wappler I, et al. The role of PI3K/AKT, MAPK/ERK and NFkappabeta signalling in the maintenance of human embryonic stem cell pluripotency and viability highlighted by transcriptional profiling and functional analysis. Hum Mol Genet. 2006;15:1894–913.
33. Huang B, Fu SJ, Fan WZ, Wang ZH, Chen ZB, Guo SJ, et al. PKCepsilon inhibits isolation and stemness of side population cells via the suppression of ABCB1 transporter and PI3K/Akt, MAPK/ERK signaling in renal cell carcinoma cell line 769P. Cancer Lett. 2016;376:148–54.
34. Hu X, Zhai Y, Kong P, Cui H, Yan T, Yang J, et al. FAT1 prevents epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway in esophageal squamous cell cancer. Cancer Lett. 2017;397:83–93.
35. Kim D, Kim S, Koh H, Yoon SO, Chung AS, Cho KS, et al. Akt/PKB promotes cancer cell invasion via increased motility and metalloproteinase production. FASEB J. 2001;15:1953–62.
|
|
|||||||||||||||||||||||||||||||||||||||||||