AbstractPurposeAlthough the mutation status of KRAS is highly concordant in primary and metastatic lesions, it has not been generalized to other major pathway genes.
Materials and MethodsIn this study, 41 genes were evaluated and the mutational profiles were compared in 46 colorectal cancer patients with paired surgical specimens of primary and metastatic lesions: synchronous (n=27) and metachronous (n=19) lesions. A high-throughput mass spectrometry-based genotyping platform validated by orthogonal chemistry, OncoMap v.4.4, was used to evaluate the formalin-fixed, paraffin-embedded surgical specimens. The patients’ demographics, tumor characteristics, and microsatellite instability status were analyzed by a retrospective chart review.
ResultsIn this study,with OncoMap, mutationswere identified in 80.4% of patientswith the following frequency: KRAS (39.1%), TP53 (28.3%), APC (28.3%), PIK3CA (6.5%), BRAF (6.5%), and NRAS (4.3%). Although 19.6% (9/46) of the patients showed no gene mutations, 43.5% (20/46) and 37.0% (17/46) had mutations in one and two or more genes, respectively. The synchronous and metachronous lesions showed similar mutational profiles. Paired samples between primary and metastatic tumors differed in 7.4% (2/27) and 10.5% (2/19) for synchronous and metachronous according to OncoMap.
IntroductionTumor progression often occurs according to the scheme of the gradual accumulation of genetic abnormalities that leads to intertumor and intratumor heterogeneity according to the spatial and temporal differences. Gerlinger et al. [1] revealed a classic example of intratumor heterogeneity with a renal cell carcinoma and its implications on tumor adaptation and therapeutic failure through Darwinian selection. These results have challenged the clinical practice of performing a single biopsy and genetic analysis of a patient, particularly in cases where the genetic profiles influence the therapeutic decisions. Therefore, the need for a clinical rebiopsy at relapse and metastasis after a resected cancer is being evaluated continuously.
Colorectal cancer (CRC) have been investigated extensively to evaluate the role of a secondary biopsy due to two factors. First, tissues from the primary and metastatic lesions have been assessable in many cases because a metastasectomy has a proven role in the care of metastatic CRC [2]. This has made it feasible to perform extensive molecular pathologic evaluations to evaluate the tumor heterogeneity within a patient. The other important factor is that RAS mutations have been important in predicting the responses to anti–epidermal growth factor receptor (EGFR) therapy [3]. Therefore, there would be a necessity to perform a second biopsy if RAS mutation status changes according to time or location with a patient. In addition, other genes that interact with RAS and affect the response to therapy can also be present.
This is the first study designed to evaluate the mutational profile of CRC lesions according to the location (primary vs. metastatic lesion) and time (synchronous vs. metachronous) in individual patients with paired specimens. This study examined whether the CRC spatial and temporal progression could be related to a difference in somatic mutations and correlate them with the clinicopathologic features.
Materials and Methods1. Patient and tumor samplesRetrospective analysis was performed on the database of Bio-Resource Center, a de-identified tissue bank linked with the Asan Biomedical Research Environment (ABLE), an anonymized electronic medical record of the Asan Medical Center, in which patients were informed before surgery that their surgical specimens might be used for research purposes and provided consent for using their archival tissues for genetic testing. Between June 2004 and June 2012, patients with histologically confirmed CRC, who underwent surgical resection of the primary tumor with an adequate amount of tissue for genomic profiling, were selected. In addition, patients with a surgical resection of a corresponding liver metastasis were included. All tumor samples were obtained from the formalin-fixed, paraffin-embedded (FFPE) tumor specimens based on an 80% cutoff for tumor sample purity. Synchronous was defined as metastatic disease at the time or within 6 months of the original diagnosis of CRC. Metachronous was defined as the absence of metastatic disease at the time of the initial diagnosis with metastatic disease developing later than 6 months of the original diagnosis. This study was approved by the Institutional Review Board (2013-0989). The patients’ demographic and tumor characteristics were reviewed.
2. DNA extraction and genotypingThe genomic DNA was extracted from 6-μm-thick slides per FFPE block and purification of genomic DNA was performed using a QIAamp DNA FFPE tissue kit (#56404, Qiagen, Hilden, Germany). Each genomic DNA sample was eluted in 50 μL of DNase- and RNase-free water, quantified using the Quant-iT PicoGreen dsDNA Assay kit (Invitrogen/Life Technologies, Grand Island, NY), and normalized to a 5 ng/μL concentration. High-throughput profiling of somatic mutations that span 471 unique mutation sites in 41 oncogenes and tumor suppressor genes was performed using OncoMap version 4.4 Core (OncoMap_v4.4C) under the Sequenom MassARRAY technology platform (Sequenom, San Diego, CA) [4,5].
3. Detection of microsatellite instability and loss of heterozygosityThe microsatellite instability (MSI) status of the tumors was determined based on the Bethesda panel (BAT25, BAT26, D5S346, D2S123, and D17S250). The polymerase chain reaction products were run on an ABI Prism 3130XL DNA Sequencer (Applied Biosystems, Foster City, CA), and analyzed using GeneScan 3.1 software (Applied Biosystems) according to the manufacturer's instructions. Tumors with two or more unstable markers were classified as MSI-high, whereas those with none or one unstable marker were classified as microsatellite stable or low level MSI. Additional loss of heterozygosity (LOH) or MSI were assayed for D18S57, D18S58, and D18S474 using the same instrument. Tumors with a 50% or more signal intensity difference between the two alleles were defined as LOH positive cases.
4. Statistical analysesThe continuous variables are presented as the medians and range. The categorical variables are presented using contingency tables and were compared using a chi-square test or Fisher exact test. The differences were considered statistically significant at p < 0.05.
The overall survival (OS) was the primary endpoint for this study and was calculated from the date of surgery until the date of death. The relapse-free survival (RFS) was calculated from the date of surgery until the date of relapse. The patients who died without relapse were censored at the time of death. The patients lost to follow-up were censored at the time of the last visit. The Kaplan-Meier method was used to estimate the OS and RFS. The survival rates were compared using a log-rank test.
Results1. Clinicopathologic characteristicsThis study reviewed 105 patients with various stages of CRC, who all received a surgical resection and had sufficient tissue of genomic profiling (Supplementary Table 1). Of those patients, this study selected a subset of 46 patients (43.8%, 46/105), who had paired samples from the primary and metastatic tumor in synchronous (n=27) and metachronous (n=19) lesions as a result of metastasectomy (Table 1). The median follow-up of these patients was 117.1 weeks (range, 39.7 to 393.6 weeks). Most (16/19) of the metachronous patients had adjuvant chemotherapy following the resection of the primary lesion. The median interval between the primary surgery and initial recurrence for the metachronous group was 83.9 weeks (range, 27.3 to 344.3 weeks). MSI and a loss of heterozygosity were obtained in 44 patients. MSI was confirmed in only one patient with a synchronous metastasis (1/44) and LOH in 24.0% (6/25) and 42.1% (8/19) for synchronous and metachronous patients, respectively.
2. Mutational profileMutations were identified in 80.4% (35/46) of the patients (Table 2), which was similar to the other patients within the Bio-Resource Center (mutation rate, 70.5%, 74/105) (Supplementary Table 2). The degree of concordance of the gene mutations between the primary and metastatic tumors were examined in both synchronous and metachronous metastatic lesions. The genomic profile of synchronous and metachronous diseases did not show a significant difference (Table 2). In addition, most of the paired samples between the primary and metastatic tumors were concordant and differed in 7.4% (2/27) and 10.5% (2/19) for synchronous and metachronous, respectively. In the synchronous patients, one patient had an APC mutation in the metastatic liver alone, while the other patient had a KRAS mutation only in the primary lesion. On the other hand, one patient had a TP53 mutation in the metastatic liver alone, whereas the other patient had a BRAF mutation only in the primary lesion in the metachronous group. Although there was one patient with MSI in the paired analysis, a total of four patients in the 105 patients were evaluated by OncoMap analysis (Supplementary Table 2). All four patients had at least one mutation: KRAS, 1/4; APC, 2/4; STK11, 0/4; TP53, 2/4; BRAF, 1/4; MLH1, 0/4; AKT1, 1/4; PIK3CA, 1/4; NRAS, 0/4; and CTNNB1, 0/4.
3. Clinical outcomeAll the metastatic diseases were resected in the 46 patients with synchronous and metachronous disease (Table 1). Recurrence was observed in 80.4% (37/46) of the patients after the metastasectomy. The median RFS was 43.0 weeks (95% confidence interval, 27.7 to 58.3) and there was no significant difference between the synchronous and metachronous disease (p=0.870) (Fig. 1). Owing to the limited follow-up, only 19.6% (9/46) had expired and the median OS could not be evaluated.
DiscussionIn this study with OncoMap, mutations were identified in 80.4% of the patients with the following frequency: KRAS (39.1%), TP53 (28.3%), APC (28.3%), PIK3CA (6.5%), BRAF (6.5%), and NRAS (4.3%). Although 19.6% (9/46) of the patients showed no gene mutations, 43.5% (20/46) and 37.0% (17/46) had mutations in one and two or more genes. The synchronous and metachronous lesions showed no difference in the mutational profiles. The paired samples between the primary and metastatic tumors differed in 8.3% (2/24) and 9.1% (2/22) for the synchronous and metachronous lesions, respectively.
The clinical impact of a rebiopsy at relapse has been controversial. One important aspect is whether the molecular information changes from the primary biopsy/resection until relapse. This may be related to the temporal and spatial differences between the primary and relapsed lesions. In addition, the therapeutic intervention between the primary and relapsed lesions, such as adjuvant chemotherapy, may also induce changes at the molecular level.
RAS mutations have been important for predicting the response to anti-EGFR therapy [3]. Whether the RAS mutation status was stable throughout disease progression and treatment is unclear. A recent meta-analysis by Mao et al. [6], who pooled studies on the concordance rates of KRAS, BRAF, and PIK3CA mutation between primary and metastatic lesion, showed that the pooled concordance rate was 92.0% for KRAS in 43 studies including 2,774 pairs of primary and metastatic lesions. In that analysis, 9% of patients with wild-type KRAS in the primary tumors who received anti-EGFR treatment had mutant KRAS in the metastases, whereas 11.3% patients with the mutant KRAS primary tumors had wild-type KRAS in the metastases. The concordance for other mutations was also high: 96.8% for BRAF, 93.9% for PIK3CA, and 71.7% for PTEN. In addition, one study evaluated the impact of the discordant cases on clinical care and found that the information obtained by additional mutation analysis changed the medical decision for subsequent therapy in 2% (6/305) of the cases [7]. On the other hand, Misale et al. [8] showed that the emergence of RAS mutations can be observed both in tissue and plasma DNA after anti-EGFR therapy. In their study, the acquisition of secondary KRAS mutations in 60% (6/10) of the cases were observed by deep sequencing (high coverage 454 sequence analysis or BEAMing) [8]. Cytotoxic agents alone, however, have not shown changes in the mutational profiles in studies including Misale et al. [8] and the present study. Because anti-EGFR therapy has no role in an adjuvant setting, the role of a rebiopsy at relapse after the standard operation and adjuvant cytotoxic chemotherapy may be limited [9].
Another important issue is the platform for sequencing. Lee et al. [10] assessed the polyclonality and genetic heterogeneity in CRC by performing targeted exome sequencing and high resolution copy number variation analysis of 15 triplets of normal colorectal tissue, primary, and matched synchronous liver metastatic lesions. In that study, the discordance rate of the KRAS mutation between the primary-metastatic pairs was 50% (3/6). On the other hand, Brannon et al. [11] used IMPACT targeted sequencing, a next generation sequencing method, for 230 key cancer-associated genes for 69 matched primary and metastatic tumors, and reported 100% concordance for the KRAS, NRAS, and BRAF mutational profiles. The present study had a discordance rate of less than 10% for the 41 genes evaluated by OncoMap in 46 matched tumors. The identification of somatic mutations using an OncoMap system was performed with two different sequential reactions, called iPLEX and homogenous Mass Extension (hME) chemistries. Screening of the mutation candidates was done with Sequenom MassARRAY iPLEX, followed by a specific hME reaction for further validation for each detected mutation. The genotypes were named using the Typer Analyzer module in the MassARRAY Typer 4 (Agena Bioscience) software, and further reviewed manually by two independent researchers to clarify uncertain calls due to clustering artifacts. The detection sensitivity of Sequenom MassARRAY technology is approximately 5%. The results obtained from the iPLEX and hME assays were compared; only the concordant calls are regarded as validated mutations. As the samples had a tumor percentage of at least 80%, the discordancy was attributed to actual sample characteristic rather than the low mutant allele frequency. Future studies and practice may differ according to platforms; therefore, the issues among platforms must be validated in precedent to make further conclusions.
Hypermutation and MSI should also be considered when interpreting the heterogeneity of mutations. Two out of three metastatic cases with discordant KRAS mutations showed hypermutation in one study [10]. The discordance may be the result of acquired mutations in the DNA mismatch repair genes after the carcinogenesis of primary tumors, which leads to dynamic polyclonal evolution. On the other hand, in the present study, all four cases with discordant mutations were microsatellite stable. As MSI is not often checked in a metastatic setting, more data will be needed to fully evaluate this issue. The clinicopathologic factors associated with MSI were difficult to characterize due to the limited number of patients (one in the synchronous group). On the other hand, four patients were identified in the expanded analysis. All patients had right side colon cancer, but there was no specific pattern in the comutations or number of mutations (KRAS, 1/4; APC, 2/4; TP53, 2/4; BRAF, 1/4; AKT1, 1/4; and PIK3CA, 1/4).
The biology of the metachronous and synchronous metastatic tumors in CRC is controversial. While some have observed no prognostic difference between the two entities, others have suggested metachronous disease to have a favorable prognosis [12,13]. Some authors have also suggested that the two entities have a different biology [14]. Rose et al. [15] evaluated KRAS and showed no difference between the two entities. This finding was expanded to 41 genes that cover the major signal pathways including the RAS–RAF–mitogen-activated protein kinase (MAPK) pathway for both primary and metastatic lesions. Although limited in size, this study showed a similar clinical outcome and mutational profiles of metachronous and synchronous metastatic tumors.
This study had several limitations. Patients who have received prior anti-EGFR therapy were not included. The use of anti-EGFR therapy and other molecular targeted agents is increasing prior to surgery in borderline resectable or conversion cases of metastatic CRC [16]. These agents may have an impact on the mutational profile, particularly the RAS-RAF-MAPK pathway. In a relapsed setting, however, most of the patients will be exposed only to cytotoxic agents due to the limited role of anti-EGFR therapy in an adjuvant setting. Second, metastatic lesions were limited to the liver. Metastatic lesions of the lymph node and lung may have different characteristics to the liver-limited lesions [17,18]. In the present clinical practice, however, the majority of metastasectomy cases will be related to liver-limited disease because its role has been established in other settings.
ConclusionThese results suggest that a conventional second biopsy or mutational profiling may not be helpful in managing CRC patients who have relapsed after adjuvant chemotherapy. In addition, metachronous and synchronous CRC has concordant mutational profiles in both primary and metastatic tumors. On the other hand, as highly reliable and sensitive mutation profiling methods, such as BEAMing, emerge and more patients are exposed to preoperative anti-EGFR therapy, this may need to be reevaluated in both tissue and liquid biopsy specimens.
Electronic Supplementary MaterialSupplementary materials are available at Cancer Research and Treatment website (http://www.e-crt.org).
AcknowledgmentsThis research was supported by a grant of the Korea Health Technology R & D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI06C0868, HI14C1061), Leading Foreign Research Institute Recruitment Program (NRF 2011-0030105 to S.J.J) through the National Research Foundation of Korea and the Asan Institute for Life Sciences, Seoul, Republic of Korea (2015-0753). The biospecimen and data used in this study was provided by the Asan Bio-Resource Center, Korea Biobank Network (2014-18 (87)).
Fig. 1.Relapse-free survival of synchronous (n=27) and metachronous (n=19) metastasis after metastasectomy. 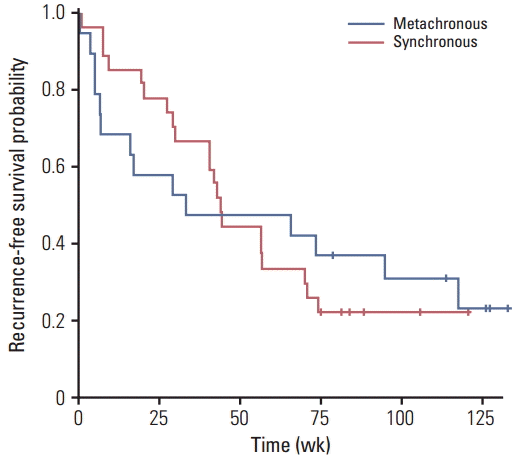
Table 1.Characteristics of the colorectal patients with paired primary and metastatic mutational profiles
Table 2.Mutational profile of the patients according to stage and paired analysis of primary and metastatic lesions References1. Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92.
2. Mahmoud N, Bullard Dunn K. Metastasectomy for stage IV colorectal cancer. Dis Colon Rectum. 2010;53:1080–92.
3. Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: can testing of tumor tissue for mutations in EGFR pathway downstream effector genes in patients with metastatic colorectal cancer improve health outcomes by guiding decisions regarding anti-EGFR therapy? Genet Med. 2013;15:517–27.
4. Wright AA, Howitt BE, Myers AP, Dahlberg SE, Palescandolo E, Van Hummelen P, et al. Oncogenic mutations in cervical cancer: genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer. 2013;119:3776–83.
5. Kim YM, Lee SW, Chun SM, Kim DY, Kim JH, Kim KR, et al. Analysis and comparison of somatic mutations in paired primary and recurrent epithelial ovarian cancer samples. PLoS One. 2014;9:e99451
6. Mao C, Wu XY, Yang ZY, Threapleton DE, Yuan JQ, Yu YY, et al. Concordant analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression between primary colorectal cancer and matched metastases. Sci Rep. 2015;5:8065.
7. Knijn N, Mekenkamp LJ, Klomp M, Vink-Borger ME, Tol J, Teerenstra S, et al. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br J Cancer. 2011;104:1020–6.
8. Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6.
9. Kawamoto Y, Tsuchihara K, Yoshino T, Ogasawara N, Kojima M, Takahashi M, et al. KRAS mutations in primary tumours and post-FOLFOX metastatic lesions in cases of colorectal cancer. Br J Cancer. 2012;107:340–4.
10. Lee SY, Haq F, Kim D, Jun C, Jo HJ, Ahn SM, et al. Comparative genomic analysis of primary and synchronous metastatic colorectal cancers. PLoS One. 2014;9:e90459
11. Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014;15:454.
12. Ghiringhelli F, Hennequin A, Drouillard A, Lepage C, Faivre J, Bouvier AM. Epidemiology and prognosis of synchronous and metachronous colon cancer metastases: a French population-based study. Dig Liver Dis. 2014;46:854–8.
13. Chu J, Goktepe O, Cheung WY. Natural history and outcomes of synchronous and metachronous colorectal cancers (CRC): a population-based analysis. J Clin Oncol. 2013;31(Suppl):Abstr 3593.
14. Slesser AA, Georgiou P, Brown G, Mudan S, Goldin R, Tekkis P. The tumour biology of synchronous and metachronous colorectal liver metastases: a systematic review. Clin Exp Metastasis. 2013;30:457–70.
15. Rose JS, Serna DS, Martin LK, Li X, Weatherby LM, AbdelMisih S, et al. Influence of KRAS mutation status in metachronous and synchronous metastatic colorectal adenocarcinoma. Cancer. 2012;118:6243–52.
16. El Zouhairi M, Charabaty A, Pishvaian MJ. Molecularly targeted therapy for metastatic colon cancer: proven treatments and promising new agents. Gastrointest Cancer Res. 2011;4:15–21.
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||